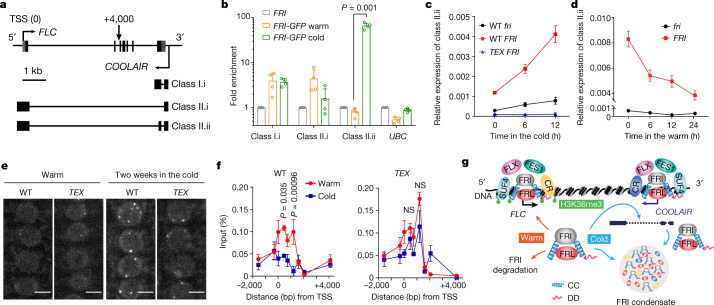
Fig. 3. COOLAIR promotes cold induction of FRI–GFP nuclear condensates and sequestration of FRI from the FLC promoter.
a, Schematic of FLC and COOLAIR transcripts at the FLC locus. Untranslated regions are indicated by grey boxes and exons by black boxes. kb, kilobase; TSS, transcription start site. b, RNA-IP assay of spliced COOLAIR enrichment by FRI–GFP with UBC as control. Mean ± s.d.; n = 4 replicates over 2 biologically independent experiments. Two-tailed t-test. c, d, Relative transcript level of COOLAIR class II.ii in the indicated plants within the same time course of changed temperatures as in Fig. 2 by RT–qPCR. Mean ± s.e.m.; n = 4 (c) and 3 (d) biologically independent experiments. e, Confocal images of wild-type and TEX root tip nuclei expressing FRI–GFP. Scale bars, 5 μm. For quantitative analysis, see Extended Data Fig. 9j, k. f, FRI–GFP occupancy on FLC promoter region in WT and TEX plants by CHIP. Mean ± s.e.m.; n = 3 biologically independent experiments. The exact distance from TSS referred to a. Two-way ANOVA adjusted by Sidak’s multiple comparisons test. NS, no significance. g, A working model for temperature-controlled FRI nuclear condensation in FLC transcriptional regulation. CC, coiled-coil domain; CR, co-transcriptional regulators; DD, disordered domain.