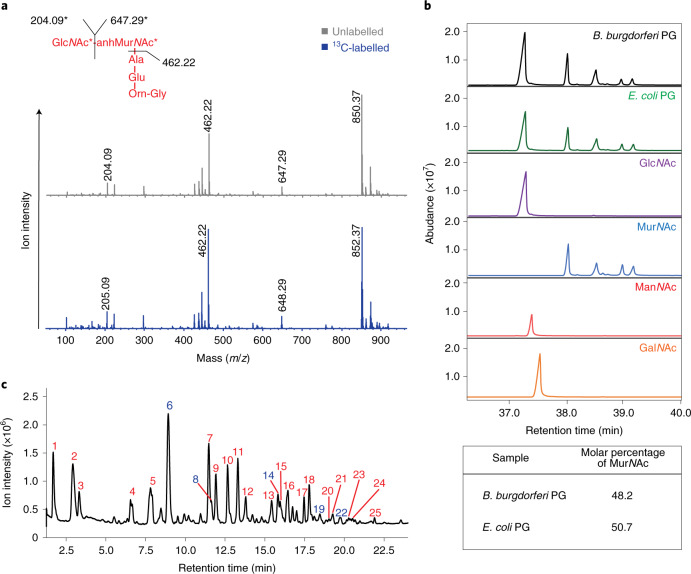
Fig. 1. Elucidating the peptidoglycan glycan strand composition of B. burgdorferi.
a, MS/MS of the GlcNAc–MurNAc–AlaGluOrnGly muropeptide from B. burgdorferi 5A11 cultured in unlabelled (grey) and [1-¹³C]ManNAc (blue), respectively. Fragmentation data confirm the location of the labelled carbon resides in the glycan component and not the stem peptide. b, Monosaccharide analysis of purified peptidoglycan isolated from B. burgdorferi 5A11 and E. coli K-12. Results were compared with reference standards GlcNAc, MurNAc, ManNAc and GalNAc (below). The inset table highlights the molar percentage of MurNAc present in each bacterial sample. c, LC–MS chromatogram of B. burgdorferi 5A11 peptidoglycan. B. burgdorferi peptidoglycan was purified, digested with mutanolysin and analysed by LC–MS. Each peak corresponds to one or more muropeptides of interest; peaks are labelled as red (GlcNAc–MurNAc muropeptides) or blue (HexNAc–GlcNAc–MurNAc muropeptides). Co-eluting peaks can be found in Supplementary Table 2.