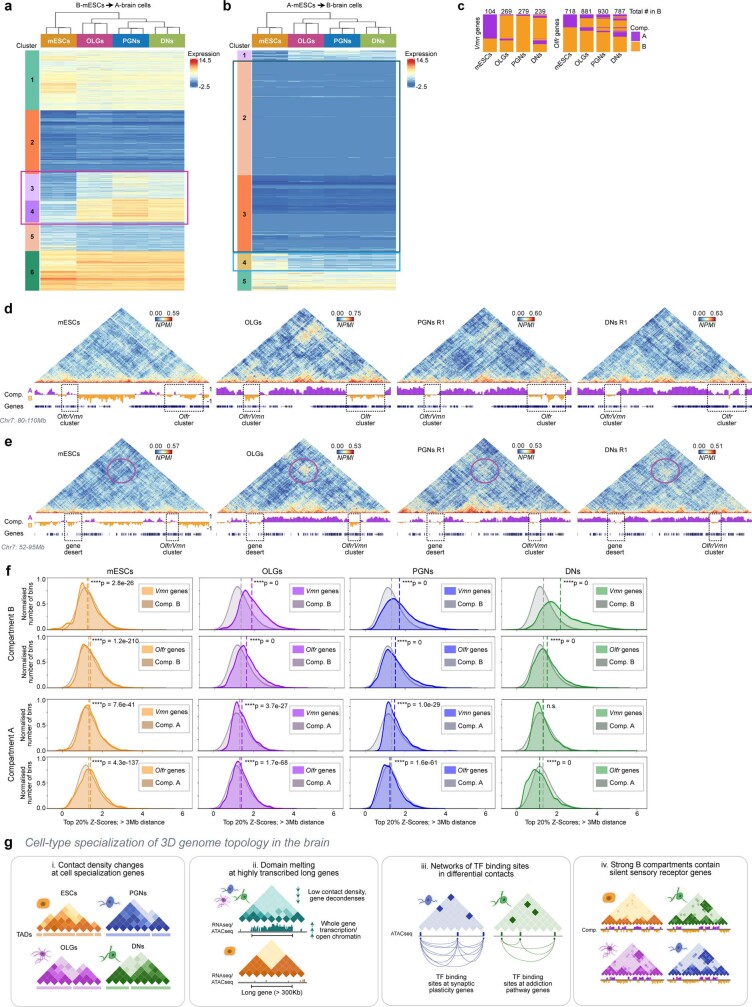
Extended Data Fig. 12. Genomic regions involved in strong long-range contacts in brain cells regions contain sensory receptor clusters in B compartments.
a, Heatmap of gene expression for genes that change compartments between compartment B in mES cells to compartment A in all brain cells. Clustering of genes by expression shows six distinct clusters where clusters 3 and 4 contain genes that increase their expression between mES cells and all brain cell types. Gene ontology (GO) in Fig. 5a was done on genes from clusters 3 and 4 combined (pink box). Expression is calculated as the R-log value for each cell type (see Methods). b, Heatmap of gene expression for genes that change from compartment A in mES cells to compartment B in brain cells. Clustering of genes by expression identifies five clusters. Genes in cluster 4 are expressed in mES cells and show lower expression in the brain cell types; they were used for GO analysis presented in Fig. 5a (light blue box). Genes in clusters 2 and 3 are not expressed in mES cells nor brain cells; they were combined and used for GO analyses presented in Fig. 5b (dark blue box). Expression is calculated as the R-log value for each cell-type. c, A higher proportion of Olfr and Vmn genes are found in B compartments in brain cells, compared to mES cells. d, GAM contact matrices show interactions between an Olfr/Vmn gene cluster and a second Olfr cluster (dashed boxes) separated by 25 Mb (Chr7: 80,000,000-110,000,000). The contacts between the two receptor clusters are strongest in OLGs, where the B compartment is strongest. e, GAM contact matrices show strong interactions that span a 30Mb distance between compartment B regions in OLGs, PGNs and DNs (purple circle), but not mES cells (Chr7: 52,000,000-95,000,000). Dashed boxes indicate contacts containing Olfr and Vmn gene clusters. f, Distribution of the top 20% of Z-Score normalized contacts for each genomic window at distances > 3 Mb (Two-sided Mann-Whitney U test; exact p-values are indicated on the plot). g, Summary diagram. The 3D genome is extensively reorganized in brain cells to reflect its gene expression specialization. (i) Contacts are rearranged at multiple scales, where formation of new TAD borders can coincide with genes important for cell specialization in all cell types. (ii) Domain melting occurs at very long genes which are highly transcribed and with high chromatin accessibility in brain cells. (iii) The most specific contacts in neurons contain complex networks of binding sites of neuron-specific transcription factors. Contacts bridge genes expressed in the neurons where the contacts are observed, with specialized functions, such as in synaptic plasticity (PGNs) and addiction (DNs). (iv) Finally, B compartments contain clusters of sensory receptor genes silent in all cell types which form strong contacts across tens of megabases.