Figure 6. CRISPR-mediated Gli3 knockdown in Axolotl causes limb skeletal condensation shift to postaxial polarity and polydactyly.
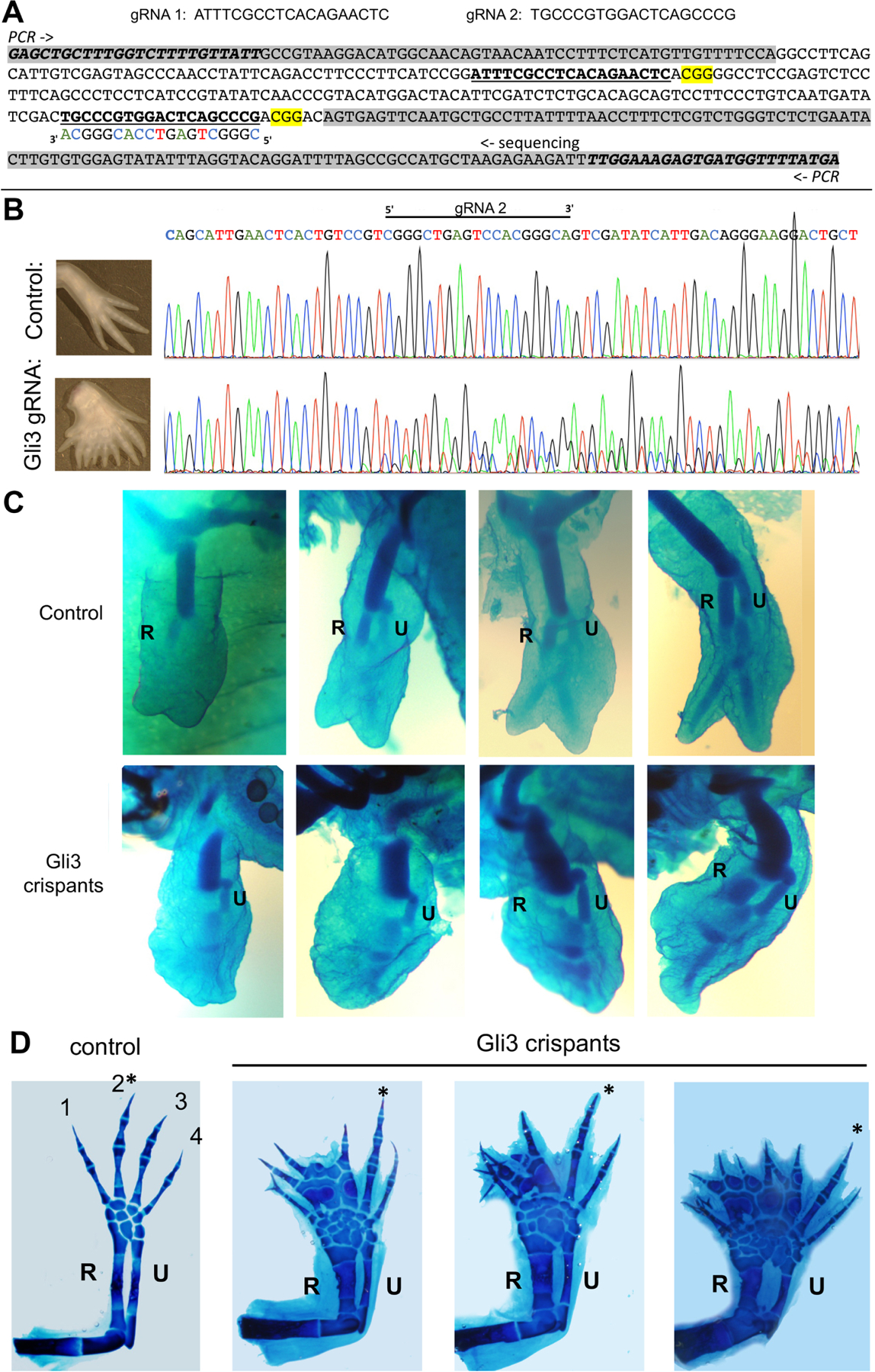
(A, B) Axolotl Gli3 guide RNAs used and sequence analysis of targeted region in crispant larvae.
(A) Top - sequences of co-injected guide RNAs targeting Axolotl Gli3 coding regions (crRNAs) together with tracrRNA and Cas9. Bottom - DNA sequence of axolotl Gli3 genomic region PCR-amplified for sequencing analyses. Intronic sequences are shaded in grey. PCR primers italicized, guides shown as bold underlined sequences, PAM sequences highlighted yellow, and complementary strand guide sequence shown as indicated in sequencing shown in B.
(B) Representative sequence trace of control compared to Gli3 crispant to verify mutagenesis (typical associated limb phenotype shown in panel to left of sequence). Direct sequencing of the PCR amplicon shows multiple nucleotide changes and sequence degeneracy following the guide/PAM region in injected founder larvae, indicative of multiple mutations.
(C) Appearance order of primary zeugopod condensations in Gli3 crispants injected with gRNAs targeting Gli3 coding regions between amino acids 176–223 (shown in Figure 6A) compared to uninjected controls ranging from approximately stage 47–50. In contrast to controls in which the anterior zeugopod (R, radius) invariably appears first (upper panels), 64% of Gli3 crispants (n=14/22, shown in lower panels) displayed postaxial dominance with posterior zeugopod (U, ulna) forming first.
(D) Mature skeletal phenotypes in Gli3 crispants compared to controls. Representative examples of preaxial polydactyly observed in 64% of Gli3 crispants (n=7/11) compared to controls (6cM, ~stage 55–57). Note that the longest digit (*) is invariably postaxial compared to controls, in which digit 2 is always largest. In addition to uninjected controls, injections of guide RNAs designed against other unrelated loci (n=500) have not resulted in any abnormal limb phenotypes. R, radius; U, ulna.
