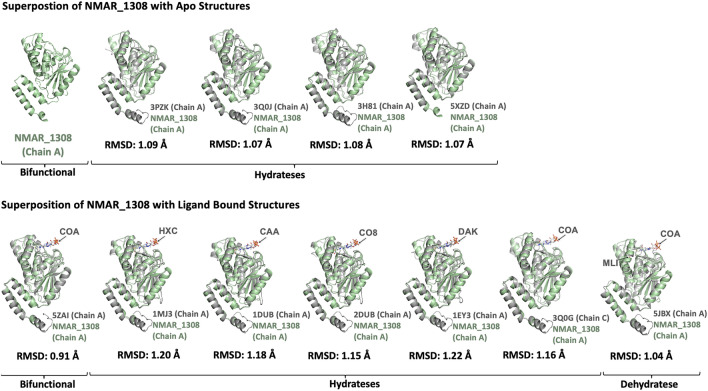
Figure 4.
Superposition of Nmar_1308 with homologous structures. Superposition is performed by using cryo-synchrotron structure of Nmar_1308 with published apo/ligand bound structures of enoyl-CoA hydrates and 3-hydroxypropionyl-CoA dehydratase (3HPD). Average RMSD values were calculated from all pairwise RMSD values between Cα atoms of an Nmar_1308 monomer and one of the superimposed monomers of homologous enzymes (36 pairs for hexamers and 18 for trimers), and are indicated below each panel. All standard errors of the RMSDs are below 0.01 Å. Chain A of Nmar_1308 is colored in pale green while the superposed structure is colored in gray. The ligands were represented with arrows. The figure is generated by PyMOL (www.schrodinger.com/pymol). (HXC: hexanoyl-CoA, CAA: acetoacetyl-CoA, CO8: octanoyl-CoA, DAK: 4-(N,N-dimethylamino)cinnamoyl-CoA, COA: coenzyme A).