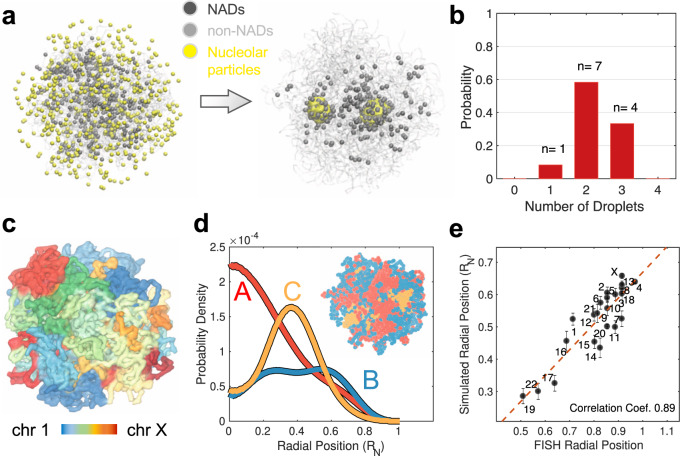
Fig. 2. Multiple droplets form in dynamical simulations of the nucleus model.
a Representative initial (left) and final (right) configurations obtained from simulations, with nucleolar particles in yellow, NADs in black, and the rest of the genome in gray. b Probability distribution of the number of droplets observed at the end of simulation trajectories. c Representative configuration of the genome that illustrates the formation of chromosome territories. d Radial distributions of the different chromatin types that support their phase separation and preferential nuclear localization. An example genome configuration is shown as the inset, with the three types colored in red (A), blue (B), and orange (C), respectively. e Simulated radial chromosome positions correlate strongly with experimental values63. Error bars correspond to the standard deviation of the 12 mean values estimated using individual simulation trajectories. Homologous chromosomes were averaged together. RN is the radius of the nucleus used in polymer simulations.