Table 1.
Inhibition data against P. aeruginosa RmlA for analogues 1a – 1f.
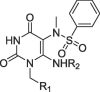
| Entry | Compd.[a] | R1 | R2 | % Inhibition at 10 μM | IC50 (μM)[b] |
|---|---|---|---|---|---|
| 1 | 1a | 4-BrC6H4 | H | 100 | 0.034 ± 0.002 |
| 2 | 1b | 4-BrC6H4 | (CH2)3NHCH3 | 100 | 0.860 ± 0.096 |
| 3 | 1c | 4-BrC6H4 | (CH2)2NHCH3 | 0 | – |
| 4 | 1d | 4-BrC6H4 | (CH2)4NH2 | 100 | 0.303 ± 0.026 |
| 5 | 1e | 4-BrC6H4 | (CH2)5NH2 | 100 | 0.316 ± 0.023 |
| 6 | 1f | Ph | 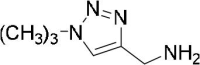 |
100 | 2.470 ± 0.020 |
[a] The following PDB codes are assigned to structures of the complexes of RmlA bound to 1b (6TQG), 1d (6T38), 1f (6T37); [b] SD, standard deviation (n = 3).
