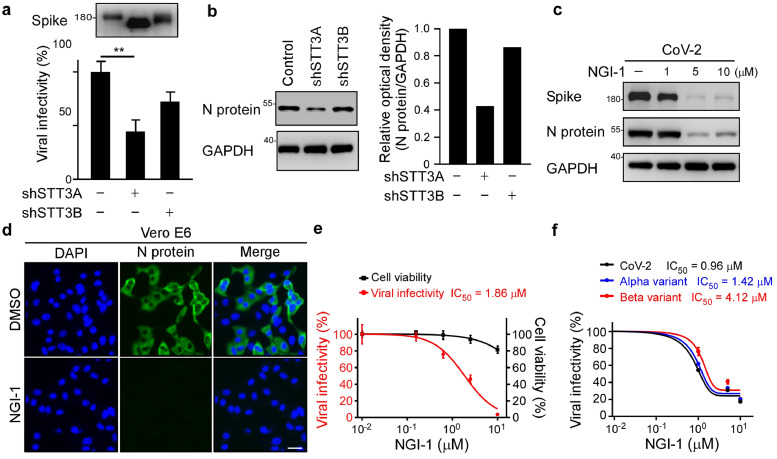
Figure 4.
Impairment of STT3A compromises SARS-CoV-2 infectivity. (a) Luciferase assay was used to monitor the infection rate of pseudovirions generated from shSTT3A or shSTT3B HEK293T cells in HEK293T-ACE2 cells. Expression of spike in pseudovirus produced from shSTT3A and shSTT3B. Statistical method: one-way ANOVA, Tukey post hoc tests. **p < 0.01 (n = 3 with mean ± SD shown). (b) Western blot analysis of N protein expression in A549-ACE2 cells infected with SARS-CoV-2 generated from control, shSTT3A or shSTT3B Vero E6 cells (left panel). N protein intensity was quantified by ImageJ (right panel). (c) Western blot analysis of spike and N protein expression in DMSO and NGI-1 treated SARS-CoV-2 infected Vero E6 cells. (d) Representative immunofluorescence images of N protein expression in DMSO and NGI-1 treated Vero E6 cell after SARS-CoV-2 infection. Scale bar, 100 μm. (e) SARS-CoV-2 neutralization IC50 of NGI-1 in Vero E6. Vero E6 cells was pretreated with NGI-1 1 h ahead at the indicated doses prior to SARS-CoV-2 infection (red). Cell viability of Vero E6 cells following NGI-1 treatment determined by MTT assay (black). Data shown are means ± SD from representative triplicates. (f) Luciferase activity was measured at 48 h to determine SARS-CoV-2, Alpha, and Beta variants pseudoviral infectivity in DMSO and NGI-1 treated HEK293T-ACE2; Data shown are means ± SD from representative triplicates.