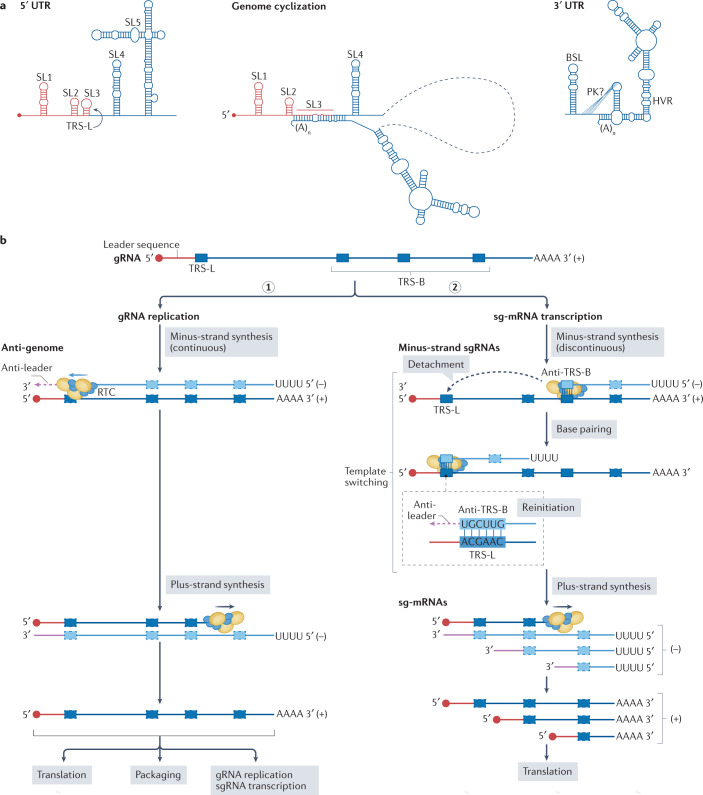
Fig. 3. SARS-CoV-2 RNA replication and transcription.
a | The genomic RNA (gRNA) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has cis-acting structures at its 5′ untranslated region (UTR) and 3′ UTR, and engages in long-distance intramolecular RNA–RNA interactions; these structures and interactions are thought to be involved in the regulation of replication and transcription. The 5′ UTR contains five conserved stem–loop (SL) structures, and the 3′ UTR contains a bulged stem–loop (BSL), a (predicted) pseudoknot (PK) and a stem–loop structure with a hypervariable region (HVR). SARS-CoV-2 gRNA cyclization results in complete opening of SL3, where the leader transcription regulatory sequence (TRS-L) resides. b | The gRNA serves as a template for gRNA replication (step 1) and for subgenomic mRNA (sg-mRNA) transcription (step 2); each process requires dedicated minus-strand templates: the anti-genome and a set of minus-strand sgRNAs, respectively. Synthesis of the latter involves a discontinuous step in which the replication–transcription complex (RTC) pauses RNA synthesis after copying one of the body transcription regulatory sequences (TRS-B), and detaches from the template. Subsequently, the RTC relocates to a position near the 5′ end of the gRNA template, where the complement of the TRS-B (anti-TRS-B) in the nascent minus-strand sgRNA engages in base pairing with the TRS-L. This template switch leads to the addition of the complement of the gRNA leader sequence (anti-leader) to the 3′ end of each of minus-strand sgRNA, which are used as templates for sg-mRNA production, thereby ensuring that all coronavirus sg-mRNAs include a 5′-terminal leader sequence of ~75 nucleotides that is identical to the 5′-terminal sequence of the gRNA. Positions of the TRSs and anti-TRSs are schematic and not drawn exactly to scale. Part b adapted from ref.161, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).