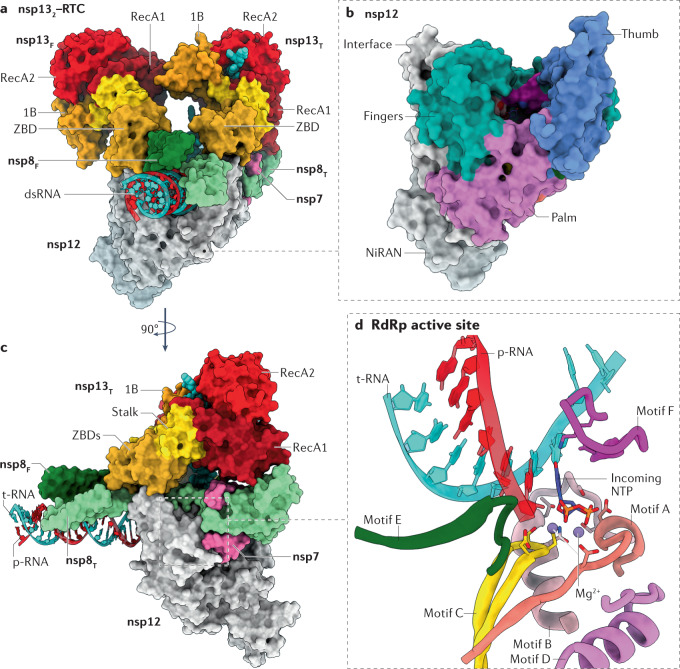
Fig. 4. Architecture of SARS-CoV-2 RNA replication and transcription complexes.
a | Surface-rendered representation illustrating two molecules of non-structural protein 13 (nsp13) bound to the replication–transcription complex (nsp132–RTC) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Depicted are the nsp7–nsp8–nsp12 RNA-dependent RNA polymerase (RdRp) holoenzyme bound to a double-stranded RNA (dsRNA) scaffold and to two molecules of nsp13: nsp13 thumb (nsp13T) and nsp13 fingers (nsp13F). The 1B, RecA1 and RecA2 domains of nsp13 are highlighted. b | The nsp12 RdRp domain adopts the canonical conformation of a ‘cupped right hand’ and its structural motifs are named accordingly. The nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain constitutes the amino-terminal 250 residues of nsp12. c | View of the nsp132–RTC complex rotated by 90° compared with part a. d | Zoomed-in view of the RdRp active site and its conserved structural motifs, except for motif G, which was excluded from the illustration for clarity. The active site is depicted with an incoming nucleotide, modelled using the pre-incorporation structures of the hepatitis C virus RdRp (Protein Data Bank entry 4WTL162). Residues involved in both chelating the catalytic Mg2+ ions and orientating the incoming nucleotide are shown in a stick representation. Protein Data Bank entry 6XEZ69 was used in preparation of this figure. NTP, nucleoside triphosphate; p-RNA, product RNA; t-RNA, template RNA; ZBD, zinc-binding domain.