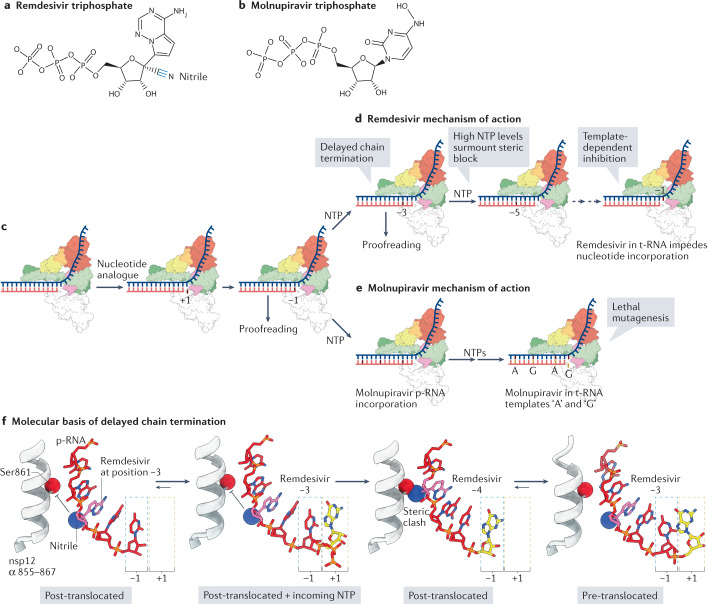
Fig. 6. Mechanisms of inhibition of coronavirus RNA synthesis by remdesivir and molnupiravir.
a | Chemical structure of remdesivir triphosphate, the activated form of remdesivir (Veklury). The ribose 1′-nitrile group is highlighted. b | Chemical structure of molnupiravir triphosphate, the activated form of molnupiravir (EIDD-2801/MK-4482). c | Schematic showing initial recognition and incorporation of a nucleotide analogue. ‘+1’ refers to the position of the nucleotide analogue before incorporation, whereas ‘−1’ refers to the position of the nucleotide analogue following catalysis and movement into the post-translocated register. d | Remdesivir exerts its inhibitory effect through ‘delayed chain termination’ and ‘template-dependent inhibition’126,134. Delayed chain termination impedes RNA-dependent RNA polymerase (RdRp) translocation through a steric clash that occurs when remdesivir reaches the fourth position (see part f) from the 3′ end of the product RNA (p-RNA) strand (red strand). Delayed chain termination is alleviated by high nucleoside trisphosphate (NTP) concentrations, leading to the proposal of template-dependent inhibition, which occurs when remdesivir is positioned at the RdRp active site in the template RNA (t-RNA) strand. e | Molnupiravir perturbs replication through ‘lethal mutagenesis’, by enabling the indiscriminate incorporation of either ATP (A) or GTP (G) when it is positioned in the t-RNA strand129,131. f | Structural analysis of the delayed chain termination mechanism. Following incorporation of remdesivir and its translocation to the −3 active site position, the ribose 1′-nitrile group is positioned to collide with the side chain of non-structural protein 12 (nsp12) Ser861 (expected clash shown by inhibitory arrows). Addition of the next nucleotide triggers the steric clash of the Ser861 side chain and the remdesivir nitrile group, forcing the growing RNA chain to populate the ‘pre-translocated’ state133. High NTP concentrations may alleviate the energetic cost imposed by the translocation barrier, by occupying the +1 site and thereby driving the forward movement of the RdRp. The structural models are based on Protein Data Bank entries 7B3B, 7B3C and 7B3D133.