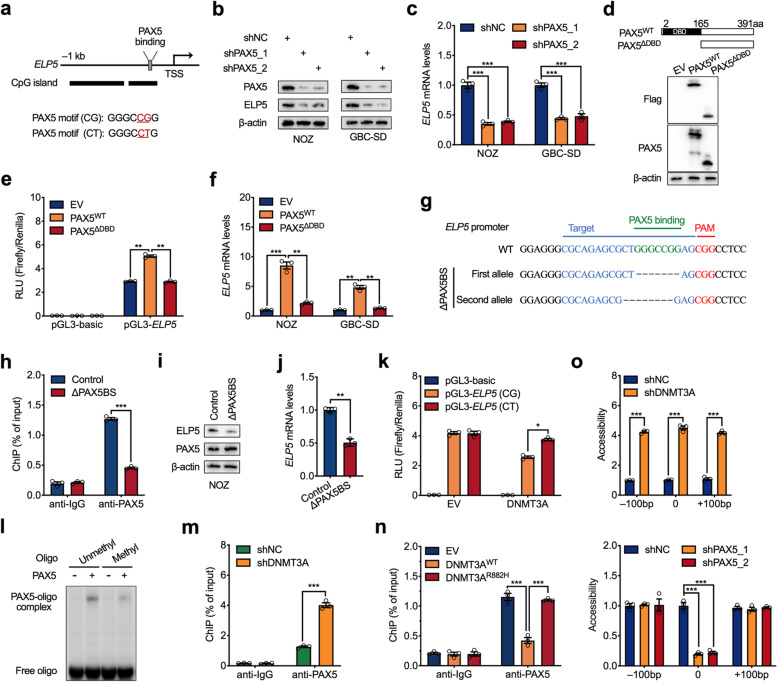
Fig. 3.
DNA hypermethylation blocks PAX5 regulation of ELP5. a Schematic diagram of PAX5 binding sites and nucleotide sequences on ELP5 promoter. b,c Western blot and RT-qPCR analysis to detect ELP5 protein levels (b) and transcription levels (c) upon PAX5 stably knockdown (shPAX5) by two independent shRNAs in NOZ and GBC-SD cells. d Schematic diagram of full length (WT) and DNA binding domain (DBD)-depleted fragment (△DBD) of PAX5 (top), and western blot to detect ectopically expressed WT and △DBD of PAX5 in HEK293T cells (bottom). e Luciferase assay of ELP5 promoter (− 450 bp to + 107 bp) activities by ELP5 promoter construct co-transfected with EV, WT, or △DBD of PAX5 construct in HEK293T cells. f RT-qPCR analysis of ELP5 transcription levels in NOZ and GBC-SD cells transfected with EV, WT, and △DBD of PAX5 constructs. g Schematic diagram showing the genomic sequences around PAX5 binding sites on ELP5 promoter in WT NOZ cell and a mutant NOZ subclone lacking the PAX5 binding site (ΔPAX5BS) generated by CRISPR/Cas9 method. h ChIP-qPCR analysis of PAX5 occupancy in ELP5 promoter in WT and ΔPAX5BS NOZ cells. i, j Western blot (i) and RT-qPCR analysis (j) to compare ELP5 expressions in WT and ΔPAX5BS NOZ cells. k Luciferase assay of ELP5 promoter (− 450 bp to + 107 bp) constructs containing methylation-sensitive (CG) or -negative (CT) PAX5 binding sites co-transfected with DNMT3A or EV in HEK293T cells. l EMSA results of PAX5 binds to the unmethylated or methylated oligonucleotides containing a PAX5 binding site in vitro. m,n ChIP-qPCR analysis of PAX5 occupancy in ELP5 promoter in DNMT3A stably knockdown NOZ cells (m) or EV, WT and R882H of DNMT3A transfected NOZ cells (n). o Chromatin accessibility is controlled by DNMT3A (top) and PAX5 (bottom) at the ELP5 promoter around PAX5 binding site in NOZ cells. The x axes indicate the distance from the PAX5 binding site. Student’s t test for statistical analysis, *P < 0.05, **P < 0.01, ***P < 0.001