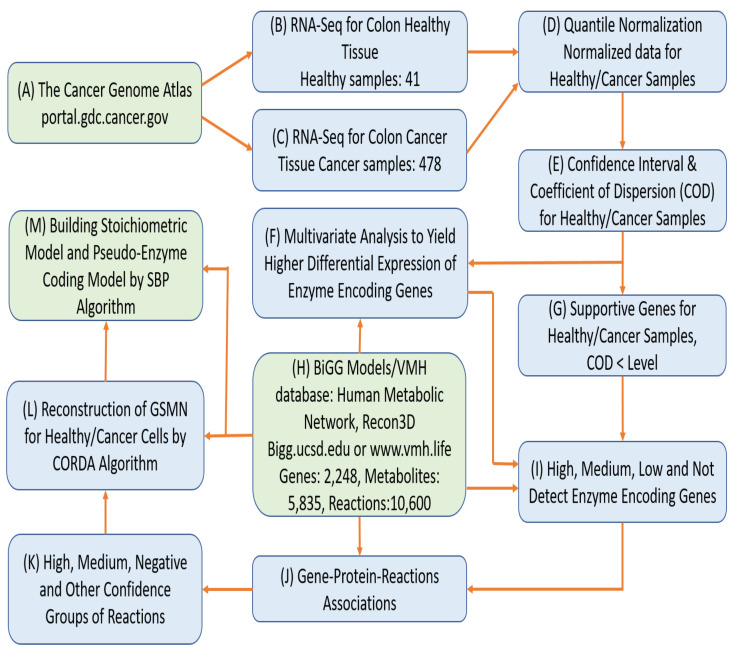
Figure 1.
Roadmap of the reconstruction of genome-scale metabolic models. Workflows for reconstructing genome-scale metabolic models for CRC and its corresponding healthy tissue. (A) Download RNA-Seq data of CRC from the TCGA database. (B–G) A series statistical analyses of the download RNA-Seq data. (H) Download general human metabolic network model (Recon3D) from Bigg Models or VMH database. (I) Integrate the Recon3D model with the RNA-Seq data to classify all enzyme-encoded genes into four classes. (J) Retrieve gene-protein-reaction associations from the Recon3D model. (K) Compute the confidence score for each reaction based on the gene-protein-reaction associations, and classify all reactions into four groups. (L) Reconstruct tissue-specific metabolic models for healthy and cancer cells using the CORDA algorithm. (M) Create GAMS codes for tissue-specific metabolic models using the SBP platform.