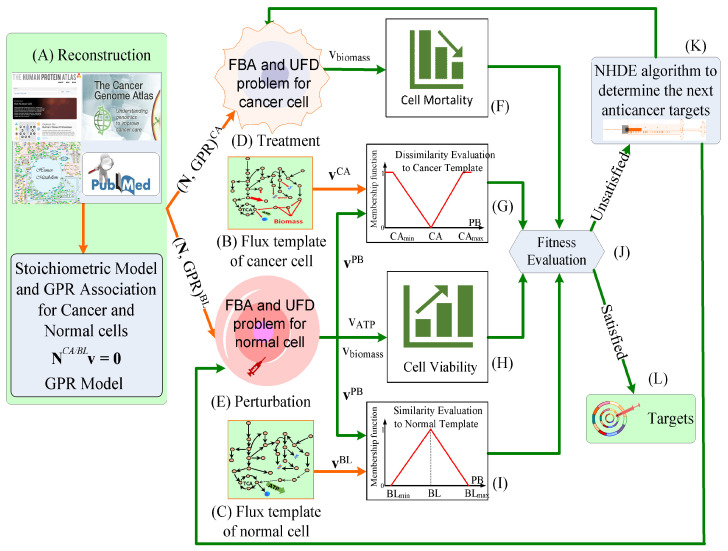
Figure 2.
Work flowchart for identifying anticancer target framework. (A) Tissue-specific genome-scale metabolic models of cancerous (CA) and normal (BL) cells were reconstructed through biological data. (B) Flux distribution patterns for cancer tissue can be provided from clinical data if available; otherwise the template can be computed through FBA and UFD problem without considered dysregulated restriction. (C) Flux distribution patterns for normal tissue can be provided from clinical data if available; otherwise the template can be computed through FBA and UFD problem without considered dysregulated restriction. (D) A set of anticancer targets are identified by the nest hybrid differential algorithm (NHDE), and provided to compute the flux distributions for each cancer treatment. (E) The same targets are provided to compute the perturbated flux distributions of normal cell during treatment. (F) Using cancer cell growth rate, cell mortality is evaluated. (G) Using membership function, cancer template and perturbated fluxes are used to compute dissimilarity grade. (H) Cell viability of perturbed cell is computed using ATP synthesis and cell growth rate. (I) Using membership function, normal template and perturbated fluxes are used to compute similarity grade. (J) The four-objective grades are used to evaluate fitness for making decision in the NHDE algorithm. (K) The next anticancer targets are generated in the NHDE algorithm if the fitness is unsatifactory, and repeat the procedures. (L) The optimal targets are obtained if the fitness is satisfactory.