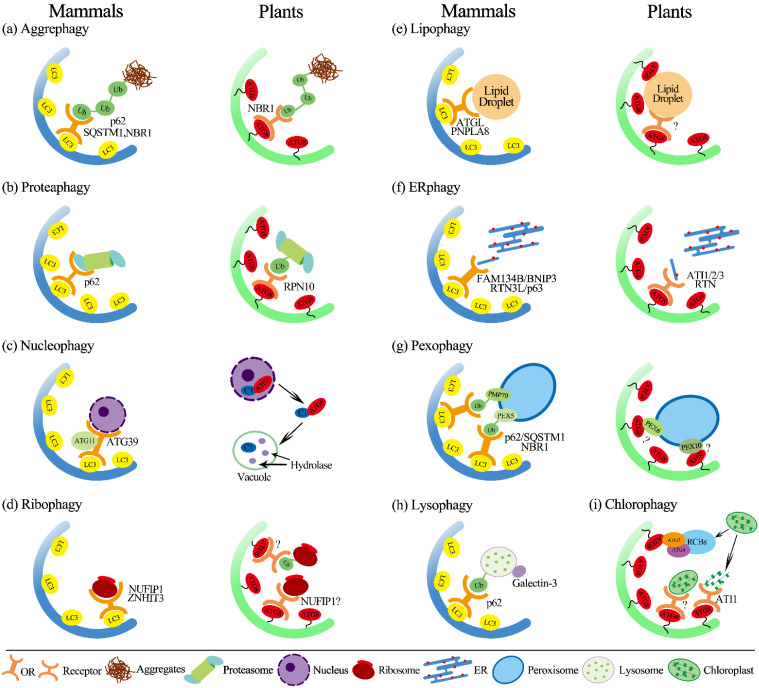
Figure 2.
Schematic representation of several mechanisms of selective autophagy in plants and animals. The degradation autophagic pathways for cell organelles and aggregates are shown and distinct features of each are highlighted. (a) Aggrephagy. Degradation of intracellular protein aggregates that form naturally or as a result of abiotic stresses that cause protein folding. Aggrephagy is activated by aggregate ubiquitylation and autophagy-binding receptors, such as NBR1 in plants and p62/NBR1 in animals. (b) Proteaphagy. Degradation of proteasomes occurs in response to proteasome inactivation or nitrogen starvation. Proteaphagy is triggered by p62 in animals and RPN10 in plants and translocates it to the cytoplasm for degradation (c) Nucleophagy. Atg39 interacts with cargo receptor Atg11 through Atg11 binding region in animals and in plants ATG8 interacts with C1 and transports it to the cytoplasm from the nucleus. (d) Ribophagy. A ribophagy receptor NUFIP1 is essential for the selective degradation of ribosomes in animals and plants. (e) Lipophagy. PNPLA8 is required to produce autophagosomes during the lipophagy process in mammals while, in plants, no receptors have been identified so far. (f) Reticulophagy. The IRE1b stress sensor is required for endoplasmic reticulum degradation, which happens in response to an accumulation of unfolded proteins during ER stress. The reticulon homology domain (RTN) containing the family of reticulophagy receptors has been identified in mammals and yeast, but not in plants. ATI1 and ATI2 were the first ER-phagy receptors discovered in plants, and FAM134B, BNIP3, RTN3, and p63 have been identified as receptors in animals that translocate it to the cytoplasm for degradation. (g) Pexophagy. Pexophagy activates in response to ROS by phosphorylating PEX5 and PMP70 leading to ubiquitination recognized by p62, targeting peroxisomes for pexophagy. No pexophagy receptors have yet been described in plants, although the LON2 chaperone likely plays a role in peroxisome stress sensing, whereas PEX6 and PEX10 interact with ATG8. (h) Lysophagy. Removal of injured lysosome via concentrated recruiting of galectin-3 and LC3 onto lysosomal membranes, as these proteins are presumably recognized by p62/SQSTM1 and targeted for degradation via autophagy. (i) Chlorophagy. Chloroplasts are degraded in a variety of ways, including piecemeal degradation of stromal fragments in Rubisco-containing bodies (RCBs) during senescence or nutrient starvation, which may be mediated by ESCRT components such as CHMP1; the engulfment of whole chloroplasts in response to oxidative damage, which may be mediated by PUB4-dependent ubiquitylation; and the formation of ATI1/2 bodies.