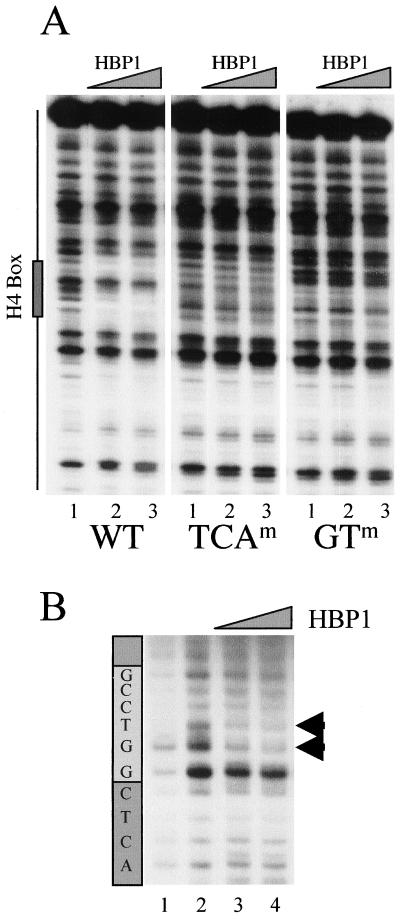
FIG. 4.
DNA-binding analysis of HBP1 on the H10 promoter. (A) A 150-bp DraIII-XbaI fragment was excised from the wild-type Xenopus H10 gene promoter, or promoters containing mutations in TCA and GT subelements (WT, TCAm, and GTm, respectively; see the legend to Fig. 2), end labeled, and incubated with 100 and 500 ng of GST-HBP1 fusion protein (lane 2 and 3 for each panel). Lane 1 shows the pattern of DNase I digestion in the absence of the protein. The position of the H4 box is indicated on the left. (B) UV laser footprinting of H4 box HBP1 complex. An end-labeled H4-box-containing oligonucleotide (32 bp) was complexed with 100 and 500 ng of purified GST-HBP1 fusion protein and irradiated with a single 266-nm laser pulse. The irradiated samples were then treated with hot piperidine, and the cleaved products were analyzed on a sequencing gel. The patterns of the cleavage of nonirradiated (lane 1) and irradiated (lane 2) naked DNA are also shown. The light gray box indicates the position of the highly conserved GGTCC motif. Arrowheads show a modified photoreactivity of the GT nucleotides in the presence of HBP1.