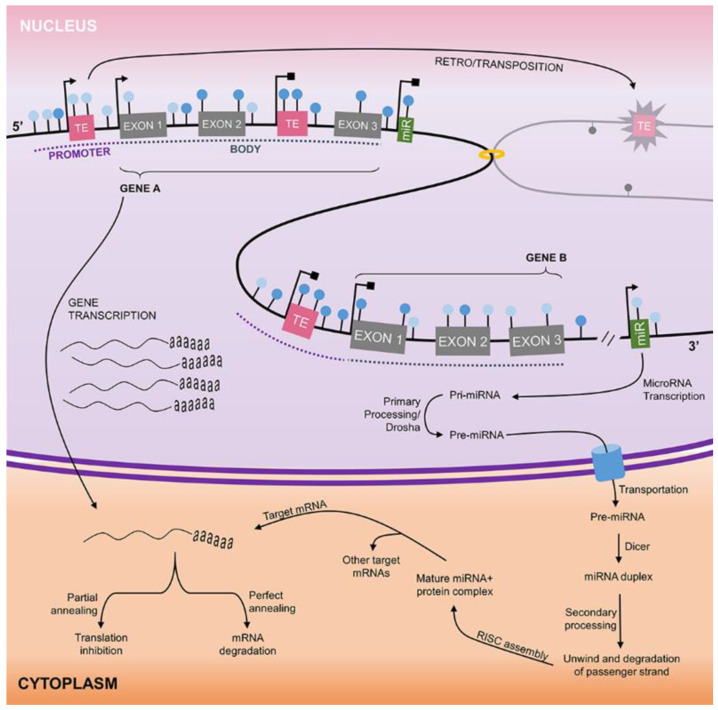
Figure 1.
Overview of gene regulation by DNA methylation and miRNA activity. Genomic DNA, found in the nucleus of eukaryotic cells, contains millions of CpG sites (lollipops) that can be either methylated (dark blue) or demethylated (light blue). These CpG sites are located in gene promoters and bodies, miRNAs, and intergenic regions, as well as in the vicinity of transposable elements (TE). When a promoter region is demethylated (Gene A), gene transcription may take place, leading to mRNA synthesis. However, when the promoter is hypermethylated (Gene B), transcription is inhibited. TEs transcription goes through the same regulatory mechanism. The transcription of some of these elements leads to the production of the transposition machinery, which enables their retrotransposition to a new genomic region, in a “copy–paste” fashion. DNA methylation similarly regulates the expression of miRNAs. Once transcribed, the primary miRNA (Pri-miRNA) is processed still in the nucleus by the RNAse Drosha, forming the precursor miRNA (Pre-miRNA). The Pre-miRNA is exported to the cytoplasm, where it is further processed by the RNAse Dicer, forming the miRNA duplex. After the miRNA duplex unwinds and the degradation of the passenger strand takes place, the lead strand (mature miRNA) assembles to the RNA-induced silencing complex (RISC) and, by nucleotide complementarity, identifies their target mRNAs. A single miRNA may have several mRNA targets and will lead to their degradation (perfect annealing) or translation inhibition (partial annealing).