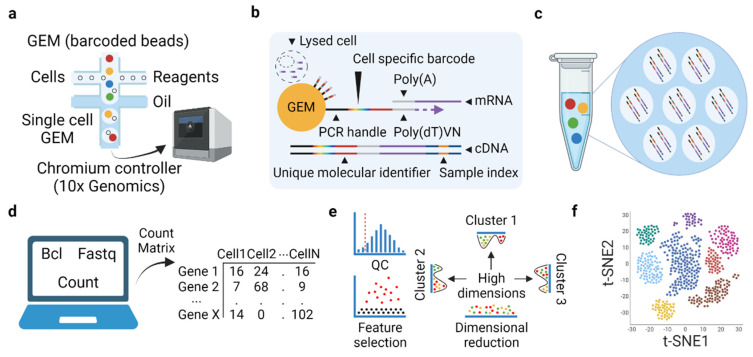
Figure 1.
Scheme of droplet-based scRNAseq and standard bioinformatics pipeline. (a) Single cells are loaded to a microfluidic system and encapsulated to an oil droplet to generate single-cell GEM. (b) RNA released from the lysed single-cell is captured by barcoded oligonucleotides and reverse transcribed to the first and second strands of DNA. (c) PCR amplifying process is conducted to generate cDNA library, which is sequenced by Illumina sequencer. (d) Cell Ranger from 10× Genomics provides raw data pre-processing pipeline, resulting in the generation of a gene expression matrix. (e) Standard pre-processing steps for scRNAseq data. Low-quality cells are removed. Highly variable features are selected and used for principal component analysis. A high-dimensional molecular profile for individual cells is computationally classified into distinct cell populations. (f) Individual cells are visualized by non-linear dimensionality reduction techniques, such as t-SNE. Abbreviation: GEM; Gel Beads-In Emulsions, t-SNE; t-distributed stochastic neighbor embedding, cDNA; complementary DNA.