Table 1.
Nomenclature of K2P-channels.
| Gene Name | IUPHAR K2P Nomenclature |
Functional Name | Other Names | Crystal Structure |
|---|---|---|---|---|
| KCNK1 | K2P1.1 | TWIK-1 (Tandem of P-domains in a weak inward-rectifying K+ channel 1) |
hOHO, DPK, KCNO1 |
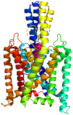
|
| KCNK2 | K2P2.1 | TREK-1 (TWIK-related K+ channel 1) |
TPKC1 |
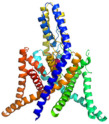
|
| KCNK3 | K2P3.1 | TASK-1 (TWIK-related acid-sensitive K+ channel 1) |
TBAK-1, OAT-1, PPH4 |
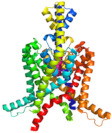
|
| KCNK4 | K2P4.1 | TRAAK (TWIK-related arachidonic acid activated K+ channel) |
FHEIG |
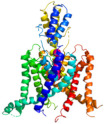
|
| KCNK5 | K2P5.1 | TASK-2 (TWIK-related acid-sensitive K+ channel 2) |
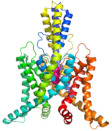
|
|
| KCNK6 | K2P6.1 | TWIK-2 (Tandem of P-domains in a weak inward-rectifying K+ channel 2) |
TOSS | - |
| KCNK7 | K2P7.1 | TWIK-3 (Tandem of P-domains in a weak inward-rectifying K+ channel 3) |
- | |
| The name kcnk8 was initially given to a murine K2P gene which was later identified as an ortholog of the human KCNK7 gene and therefore renamed to kcnk7 | ||||
| KCNK9 | K2P9.1 | TASK-3 (TWIK-related acid-sensitive K+ channel 3) |
KT3.2, BIBARS, TASK32 | - |
| KCNK10 | K2P10.1 | TREK-2 (TWIK-related K+ channel 2) |
PPP1R97 |
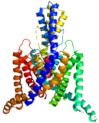
|
| KCNK11 was withdrawn due to nomenclature duplications with KCNK15 | ||||
| KCNK12 | K2P12.1 | THIK-2 (Tandem pore domain halothane inhibited K+ channel 2) |
- | |
| KCNK13 | K2P13.1 | THIK-1 (Tandem pore domain halothane inhibited K+ channel 1) |
- | |
| KCNK14 was withdrawn due to nomenclature duplications with KCNK15 | ||||
| KCNK15 | K2P15.1 | TASK-5 (TWIK-related acid-sensitive K+ channel 5) |
KT3.3, dJ781B1.1 | - |
| KCNK16 | K2P16.1 | TALK-1 (TWIK-related alkaline pH-activated K+ channel 1) |
- | |
| KCNK17 | K2P17.1 | TALK-2 (TWIK-related alkaline pH-activated K+ channel 2) |
TASK-4 | - |
| KCNK18 | K2P18.1 | TRESK (TWIK-related spinal cord K+ channel 1) |
MGR13, TRIK, TRESK2 | - |
IUPHAR, International Union of Basic and Clinical Pharmacology. Visualizations of the channel structurs were generated with PyMOL (TM) Molecular Graphics System, Version 2.3.0 (Schrodinger, LLC; New York, NY, USA) from crystall stuctures with the protein database enty numbers: 3UKM, 4TWK, 6RV2, 3UM7, 6WLV, 3UX0 and 4BW5.
