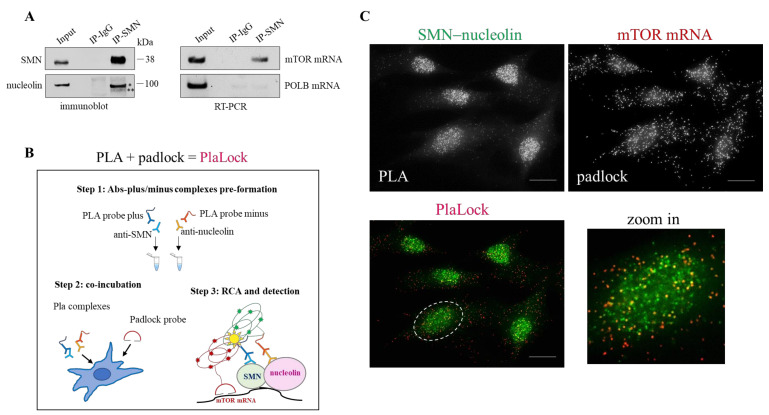
Figure 3.
SMN coexists with nucleolin–mTOR mRNA complexes. (A) Cellular extracts from hTert-Fibroblasts were subjected to RNA-immunoprecipitation (RIP) assay. SMN monoclonal antibody-conjugated beads or mouse IgG-conjugated beads were used in immunoprecipitation steps. (left) Immunoblotting validating nucleolin co-precipitation with SMN. To note, anti-nucleolin antibody detects two bands: a main band of 100 kDa (*) and a faint, smaller band (**). (right) mTOR mRNA were checked in RIP samples by RT-PCR analysed by agarose gel electrophoresis. DNA Polymerase Beta (POLB) mRNA was monitored as control. Representative panels of n = 3 independent experiments. (B) Diagram illustrating the main experimental steps of the “PlaLock” assay in fixed cells. PlaLock assay was designed to combine the PLA and padlock methods in situ. In this way, it is possible to visualize the coexistence, at subcellular levels, of a selected mRNA with a specific protein–protein complex (see Methods section). (C) Representative images of a PlaLock performed in hTert-Fibroblasts. (right) SMN-nucleolin complexes visualized by PLA. (left) Subcellular distribution of mTOR mRNA by padlock. The bottom panel visualizes the merged images. mTOR mRNA and SMN-nucleolin complexes are labelled in red and green, respectively. Yellow dots are mostly detectable in nuclear regions, as illustrated in the higher magnification of the white dotted circle region (zoom in). Scale bar 5 µm.