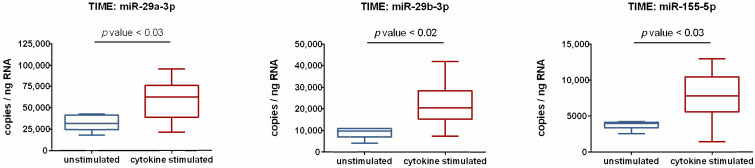
Figure 3.
Human endothelial cells (cell line TIME, ATCC® number CRL-4025) were evaluated following stimulation with 5 ng/mL each of IL-1β, TNF-α, and IFN-γ for 24 h, and compared to unstimulated controls. Absolute quantification of miRNAs from isolated total RNA was performed using ddPCR analysis. The QX 200 Droplet Digital PCR System (Bio-Rad) and specific LNA PCR primers (Qiagen) were used according to the manufacturer’s instructions. Based on the number of measured positive droplets, the system calculates the copy number of the target RNA in the total mixture, assuming a Poisson distribution. Since a defined amount of RNA sample is used in each ddPCR assay, the copy number per ng RNA can be determined. The box plots depict the median of determined miRNA expression levels, the lower and upper quantile, and the two extreme values. Each group included six biological replicates and two technical replicates. An unpaired t-test was used to identify significant differences. Statistical analysis was performed utilizing GraphPad Prism 9 (GaphPad Software, La Jolla, CA, USA). In all cases, p < 0.05 was assumed to indicate significant differences.