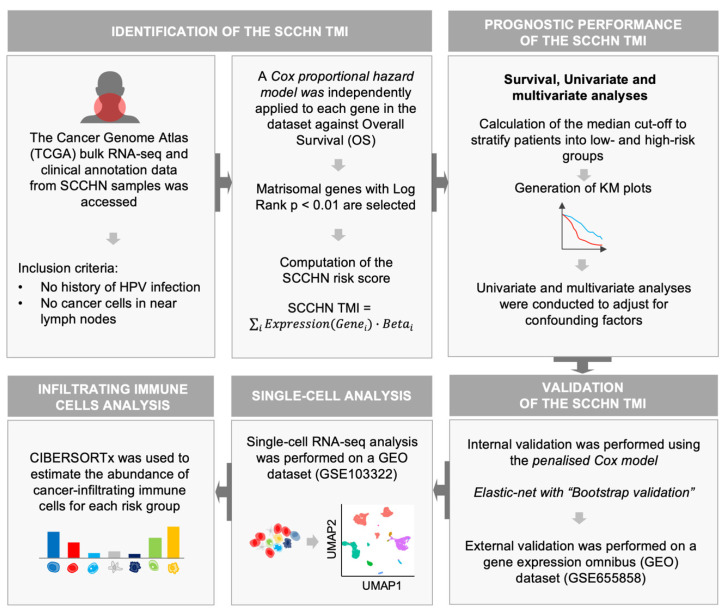
Figure 1.
Schematic illustration of the bioinformatics workflow. We first constructed a matrisome-derived SCCHN-specific signature from primary tissue samples using a bioinformatic-based approach. Specifically, access and retrieval of large datasets from “The Cancer Genome Atlas database” was conducted via XENA browser. SCCHN-tumor matrisome index (SCCHN TMI) was computed using R: (1) prognostic genes were identified through the application of the Cox proportional hazard model; (2) the median cut-off was used to stratify patients into low and high-risk and generate Kaplan−Meier (KM) plots for each risk group; (3) internal validation and external validation of the SCCHN TMI were conducted on a microarray and scRNA-seq datasets. The web-based tool CIBERSORTx was used to estimate the relative fraction of cancer-infiltrating immune cells based on the RNA-seq data, as a function of the SCCHN TMI risk group.