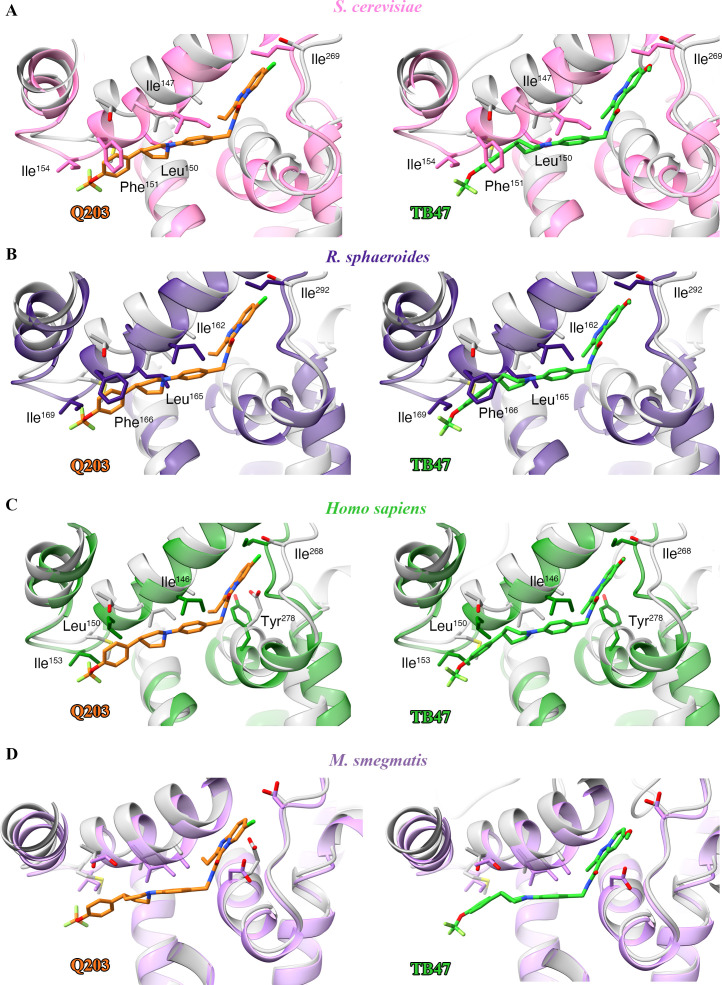
Figure 6. Structural alignment between the M. tuberculosis Qp-binding pocket where Q203 or TB47 binds with homologous subunits from four other species.
These subunits are from (A) S. cerevisiae (pink, PDB: 1KYO), (B) R. sphaeroides (blue, PDB: 2QJP), (C) Homo sapiens (green; PDB: 5XTE), and (D) M. smegmatis (violet, PDB: 6ADQ). Residues causing steric clashes in the homologous subunits are labeled.
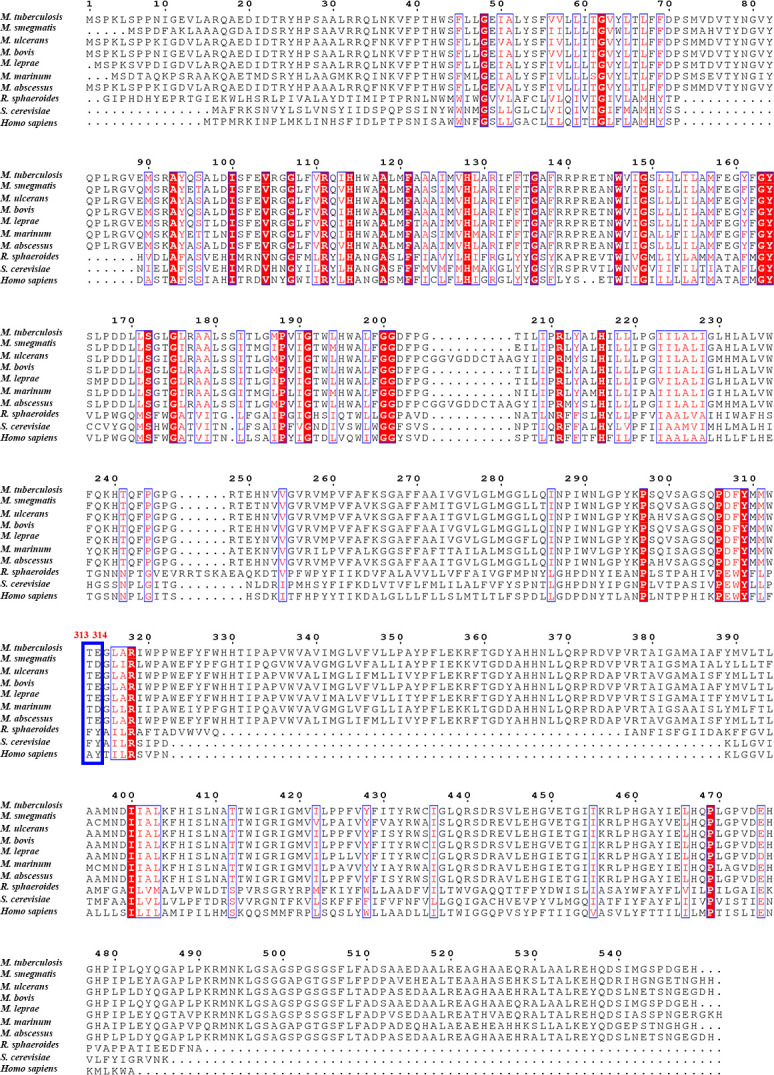
Figure 6—figure supplement 1. Sequence alignment of M. tuberculosis QcrB with their counterparts in other species including Homo sapiens.