Figure 7.
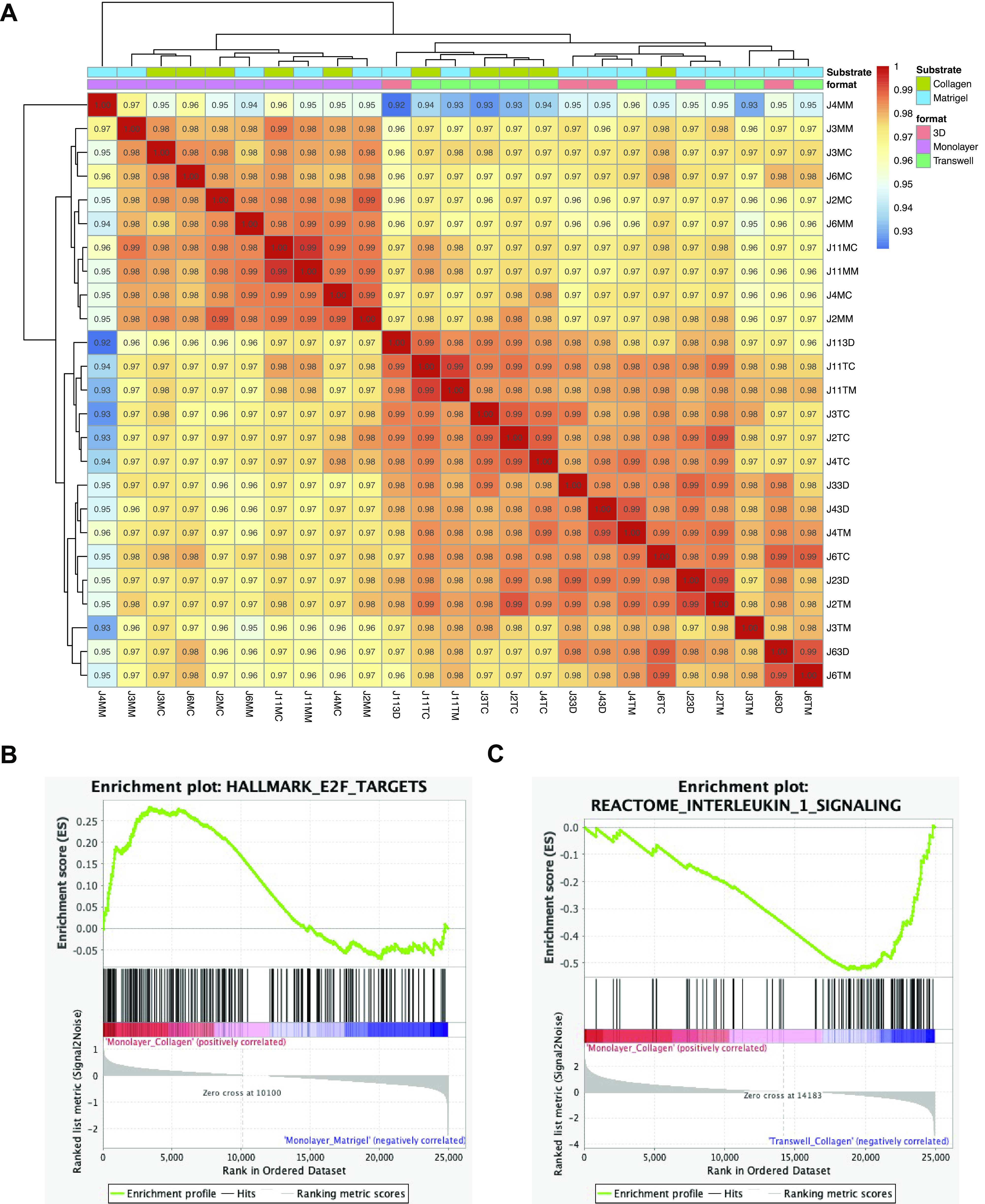
Format validated as a strong driver of variance in a synchronized experiment with control jejunal enteroids. A: validation analysis of three-dimensional (3-D), monolayer (on collagen and Matrigel) and transwell (on collagen and Matrigel) differentiated jejunal enteroids grown simultaneously with five jejunal lines (J2, J3, J4, J6, and J11) for each substrate-format combination (n = 25). B: gene set enrichment analysis (GSEA) showing an enrichment of the hallmark E2F targets gene set signature in jejunal (validation experiment) transwells grown on collagen (normalized enrichment score, NES = 1.36 and false discovery rate, FDR = 0.08) when compared with jejunal (validation experiment) transwells grown on Matrigel. C: GSEA showing an enrichment of the reactome interleukin-1 signaling gene set signature in jejunal (validation experiment) transwells grown on collagen (NES = −2.03 and FDR = 0.003) when compared with jejunal (validation experiment) monolayers grown on collagen. Correlation coefficients of the matrix were generated using the entire transcriptome.
