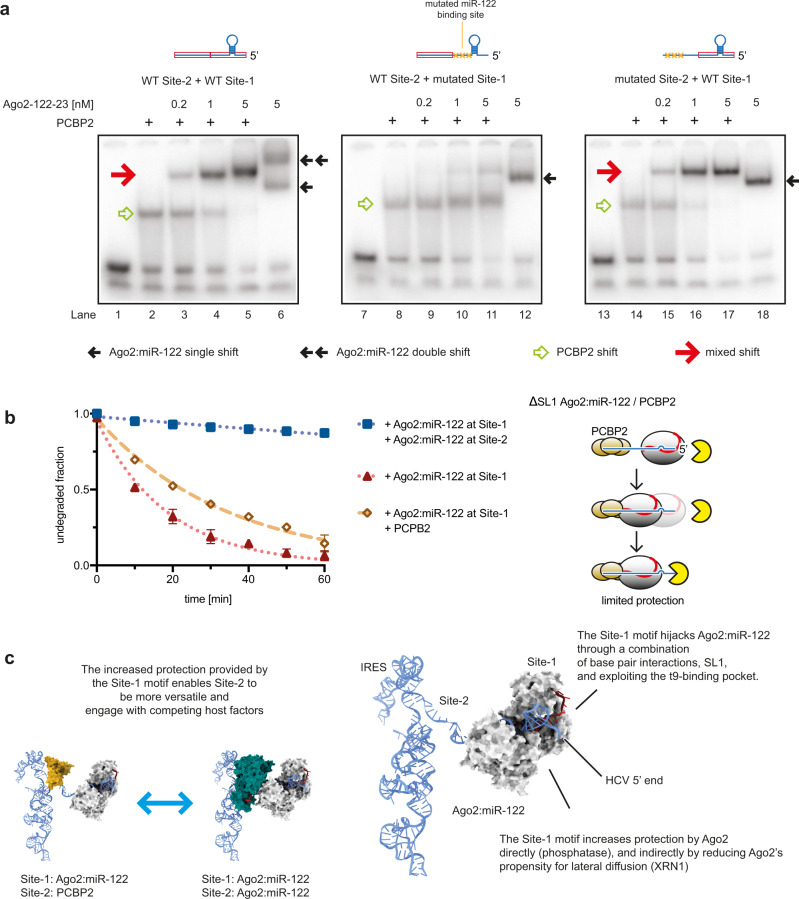
Fig. 5. A mixed Ago2:miR-122:PCBP2 complex can form on the HCV 5’ tail.
a EMSA of HCV 5’ tail RNAs by PCBP2 and/or Ago2:miR-122. WT, MUT1, and MUT2 RNAs were incubated with PCBP2 (100 nM), and/or Ago2:miR-122 (concentrations indicated), or both prior to running the gel. Lanes 1, 7, and 13 show free RNA for WT, MUT1, or MUT2. Schematic of HCV 5’ tail RNAs (blue) with miR-122 binding sites as red boxes and mutated sites indicated by orange X’s, show at the top. The experiment was repeated twice and a representative gel is shown. b XRN1 mediated degradation of Site-1 ∆SL1 MUT2 RNA in presence of Ago2:miR-122-wt, Ago2:miR-122-wt and PCBP2, or Ago2:miR-122-wt and -mut. Data shown as means of 3 independent replicates with error bars representing SEM. The cartoon (right) describes the proposed mechanism, with PCBP2 occupancy at Site-2 only partially reducing Ago2:miR-122 lateral diffusion from Site-1. c Model of Ago2:miR-122 bound to Site-1 with HCV IRES shown for scale (PDBID: 5a2q). Ago2 occupancy at Site-1 may enable a molecular switch in which Site-2 occupancy alternates between Ago2:miR-122 and PCBP2 (represented as 3 KH domains, PDBID: 2p2r).