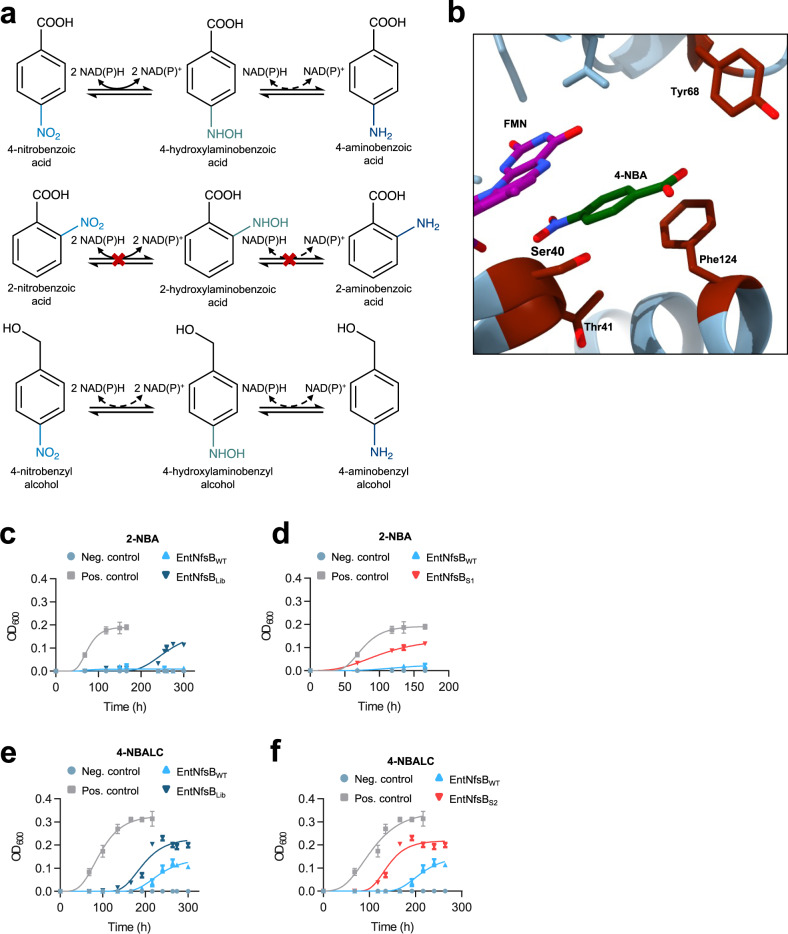
Fig. 5. Chemically-directed selection of nitroreductase variants with altered substrate specificity.
a Catalytic activities of EntNfsB (Enterobacter cloacae nitroreductase NfsB). EntNfsB is able to reduce several nitroaromatic compounds, such as 4-nitrobenzoic acid (4-NBA), using NADH or NADPH as cofactors. EntNfsBWT is also able to reduce 4-nitrobenzyl alcohol (4-NBALC) less efficiently, but does not display any activity towards 2-nitrobenzoic acid (2-NBA). b Substrate binding site of EntNfsBWT with 4-NBA bound (PDB code 5J8G). Residues 40, 41, 68, and 124 were close to the substrate, and did not contact the FMN group essential for catalysis. Anaerobic culture of AL cells transformed with the library EntNfsBLib (pLS169) in M9 glucose medium with 2-NBA (c) or 4-NBALC (e). Controls are as in Fig. 1. d Anaerobic culture of AL cells transformed with EntNfsBS1 (with substitutions S40A, T41I, and F124A; pLS169_1), the variant with activity towards 2-NBA, in M9 glucose medium supplemented with 2-NBA. f Anaerobic culture of AL cells transformed with EntNfsBS2 (with substitutions T41L, Y68L, and F124L; pLS169_3), the variant with improved activity towards 4-NBALC, in M9 glucose medium supplemented with 4-NBALC. Data points of growth curves represent mean values, with error bars showing standard deviation; n = 3 biologically independent cultures for all timepoints of growth curves. Source data are provided as a Source Data file.