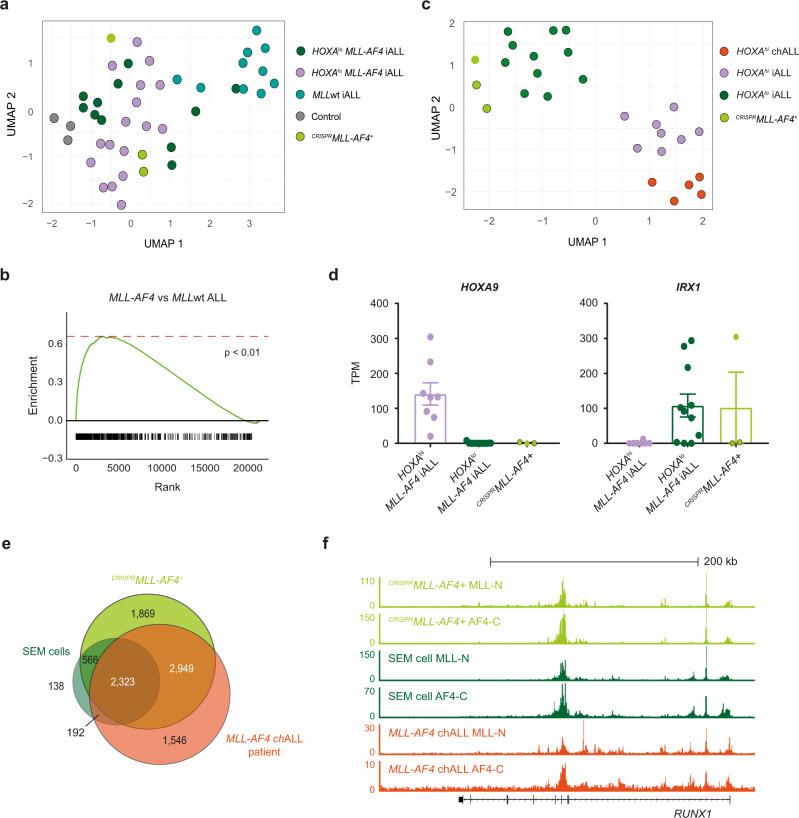
Fig. 4. CRISPRMLL-AF4+ ALL recapitulates the molecular profile of MLL-AF4 ALL in patients.
a UMAP showing clustering of CD19+ cells from CRISPRMLL-AF4+ (light green; black border = proB ALL, no border = preB-ALL) and control (gray) mice with HOXAlo MLL-AF4 (dark green, n = 11), HOXAhi MLL-AF4 (purple, n = 20), and MLLwt (blue, n = 9) infant-ALL patient samples from a publicly available dataset27 based on 7041 significantly differentially expressed genes (FDR < 0.05, edgeR glm test) between CRISPRMLL-AF4+ ALL, controls, MLL-AF4 infant-ALL, and MLLwt infant-ALL. b Gene set enrichment analysis (GSEA)52 showing CRISPRMLL-AF4+ ALL is more enriched for genes that are upregulated in MLL-AF4 ALL compared to MLLwt ALL (1000 genes) when compared to CD19+ cells from control mice (p < 0.01). c UMAP showing clustering of CRISPRMLL-AF4+ (light green; black border = proB ALL, no border = preB-ALL) with HOXAlo MLL-AF4 (dark green, n = 11) HOXAhi MLL-AF4 infant-ALL (purple, n = 8), and HOXAhi MLL-AF4 childhood-ALL (orange, n = 5) from a publicly available patient dataset23 based on 765 significantly differentially expressed genes (FDR < 0.05; edgeR glm test) between these three patient subsets (Supplementary Data 3). d Expression (TPM) of HOXA9 and IRX1 in HOXAhi MLL-AF4 infant-ALL (purple, n = 8), HOXAlo MLL-AF4 infant-ALL (dark green, n = 11) and CRISPRMLL-AF4+ ALL (light green; black border = proB ALL, no border = preB ALL). Data are shown as mean ± SEM. Source data are provided as a Source Data file. e Venn diagram showing overlap of MLL-AF4-bound genes (genes with an MLL-AF4 peak in the gene body) in CRISPRMLL-AF4+ ALL in a primary recipient mouse, the MLL-AF4+ SEM cell line and a primary MLL-AF4 childhood-ALL (chALL) patient sample. MLL-AF4 peaks = directly overlapping MLL-N and AF4-C peaks. f Representative ChIP-seq tracks at the MLL-AF4 target gene, RUNX1 in CRISPRMLL-AF4 + ALL, the SEM cell line and a primary MLL-AF4 childhood-ALL (chALL) patient sample.