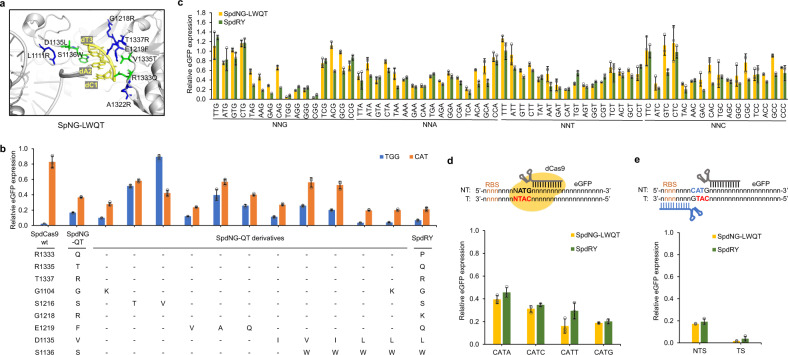
Fig. 2. Improving the activity of SpdNG-QT toward 5′-CAT-3′ PAM by mutating PAM-proximal residues.
a PAM-proximal residues involved in PAM recognition. Modeled structure of SpNG-LWQT (D1135L/S1136W/R1333Q/V1335T, residues shown in green) with 5′-CAT-3′ PAM based on SpNG (PDB ID: 6AI6). b Impact of mutating PAM-proximal residues on 5′-TGG-3′ (blue) and 5′-CAT-3′ (orange) PAM recognition. c PAM profile comparison between SpdNG-LWQT (gold) and SpdRY (green) against all NNN PAM libraries. d The influence of the fourth-position base on 5′-CAT-3′ PAM recognition of SpdNG-LWQT (gold) and SpdRY (green). e The repression effects of SpNG-LWQT (gold) and SpdRY (green) when targeting 5′-CAT-3′ PAM on both nontemplate and template strand. NTS nontemplate strand, TS template strand, RBS ribosome binding site. Data indicated the mean ± standard deviation (n = 3 independent biological replicates). Source data are provided as a Source Data file.