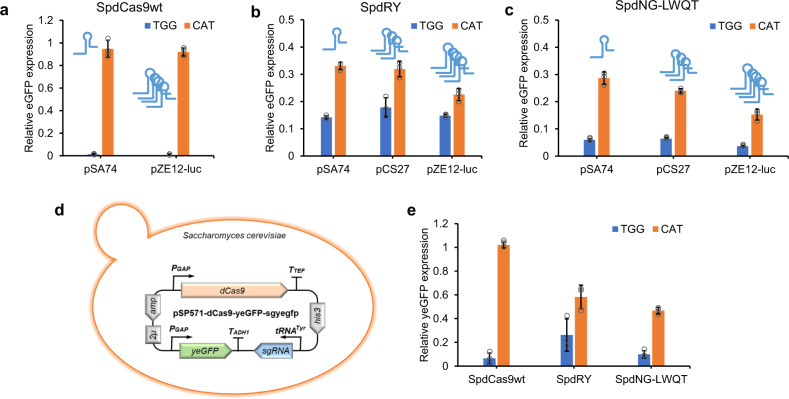
Fig. 5. Transcriptional repression of eGFP in E. coli and S. cerevisiae.
Repression of chromosomal eGFP by SpdCas9wt (a), SpdRY (b) and SpdNG-LWQT (c) targeting both 5′-TGG-3′ (blue) and 5′-CAT-3′ (orange) PAMs in E. coli BW25113(F′)::eGFP. The dCas9s were expressed on pCS27, and sgRNAs were expressed on low-copy (pSA74), medium-copy (pCS27), or high-copy (pZE12-luc) plasmids. d Construction of pSP571-dCas9-yeGFP-sgyegfp plasmid for gene repression assay in S. cerevisiae. The dCas9s and the yeast eGFP (yeGFP) were driven by the PGAP promoter, and sgRNAs were driven by the tRNATyr promoter. e Comparison of yeGFP repression by SpdCas9wt, SpdRY and SpNG-LWQT targeting both 5′-TGG-3′ (blue) and 5′-CAT-3′ (orange) PAMs in S. cerevisiae. Data indicated the mean ± standard deviation (n = 3 independent biological replicates). Source data are provided as a Source Data file.