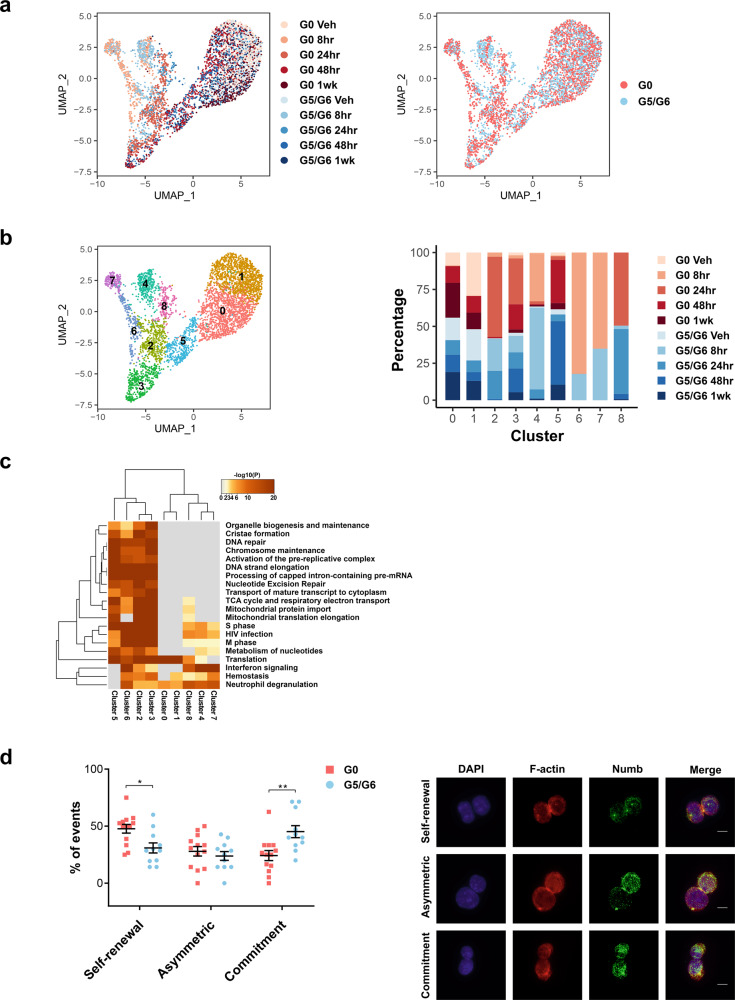
Fig. 3. HSCs with short telomeres do not undergo apoptosis, autophagy, or senescence upon IFN signaling activation.
a UMAP of scRNA-seq data displaying 2601 and 1886 pooled single G0 or G5/G6 HSCs, respectively, isolated from mice at several time points following pI:pC injection (n ≥ 8 mice per group). Each dot represents one cell. Different colors represent the sample treatments and identities (left) or identities only (right). Veh vehicle. b UMAP of scRNA-seq data from Fig. 3a showing the cell cluster distribution independently of cell identity. Each color represents a different cell cluster. Right, the distribution of G0 and G5/G6 HSCs from mice at different treatment time points among the nine scRNA-seq clusters, represented as the percentage of cells from each group that belong to each cluster. Veh vehicle. c Joint pathway enrichment analyses of the marker genes that define each of the clusters shown in Fig. 3b and Supplementary Dataset 7 (adjusted P ≤ 0.05). The Reactome gene sets are shown. d Left, frequencies of the cell division type in G0 and G5/G6 HSCs induced to proliferate in vitro. Each dot represents the mean of the frequency of the event type in pooled HSCs from ≥2 mice per pool (n = 133 and n = 113 events from 13 G0 HSC pools and 11 G5/G6 HSC pools, respectively). Bars represent the means ± S.E.M. Statistically significant differences were detected using two-way ANOVA. *P < 0.05, **P < 0.01, Asymmetric division: P = 0.87. Right, representative anti–F-actin and anti-Numb immunofluorescence in G0 HSCs undergoing the three different types of cell division. Red indicates F-actin; green, Numb; blue, DAPI. Scale bars represent 10 μm. Source data are provided as a Source Data file.