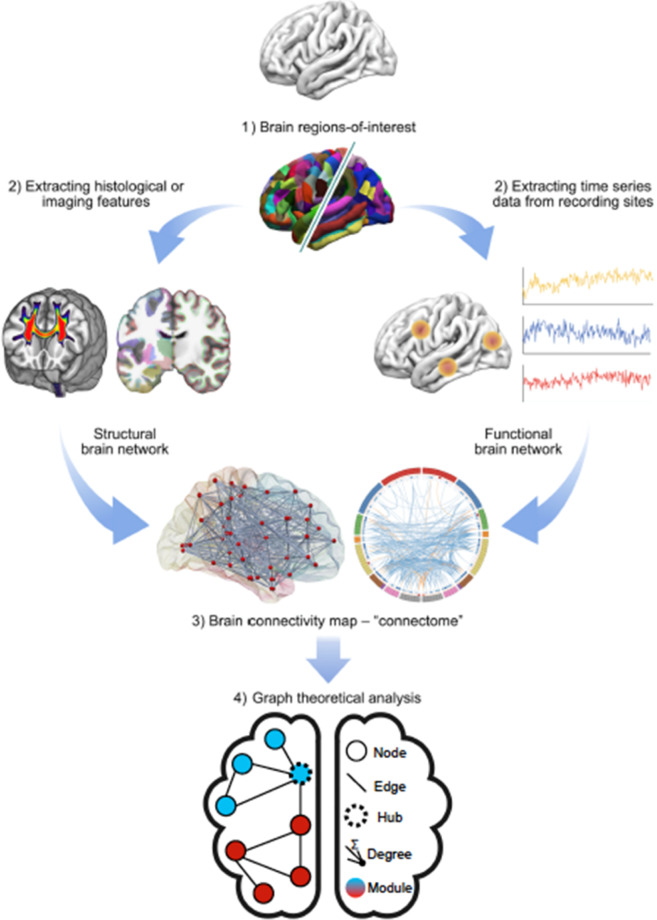
Fig. 4. Human brain network analysis: high-level schematic.
Brain regions or nodes are defined based on anatomical, functional, or multimodal parcellations, thus subdividing the whole brain into p regional nodes (1). Connectivity between nodes can be estimated in many different ways, to define the weight of an edge or connection between each possible pair of nodes. Anatomical connectivity (2, left) can be estimated by dMRI tractography, structural covariance, or morphometric similarity; functional connectivity (2, right) can be estimated by the correlation between nodal mean time series. The resulting (p × p) matrix is the connectome, which can be represented in diverse formats, including (left) an anatomical rendering, where the nodes are located at the centroid of each region and a line is drawn between nodes if their pairwise connectivity exceeds an arbitrary threshold; or (right) a ring diagram, where the regional nodes are arranged around the perimeter of the circle (color-coded according to anatomical criteria or modular affiliation) and edges traverse the interior of the circle denoting suprathreshold connectivity. Finally, the complex topological properties of these networks, including hubs and modules, can be estimated using tools from graph theory, here illustrated for the simplest class of binary undirected graphs (from [236]).