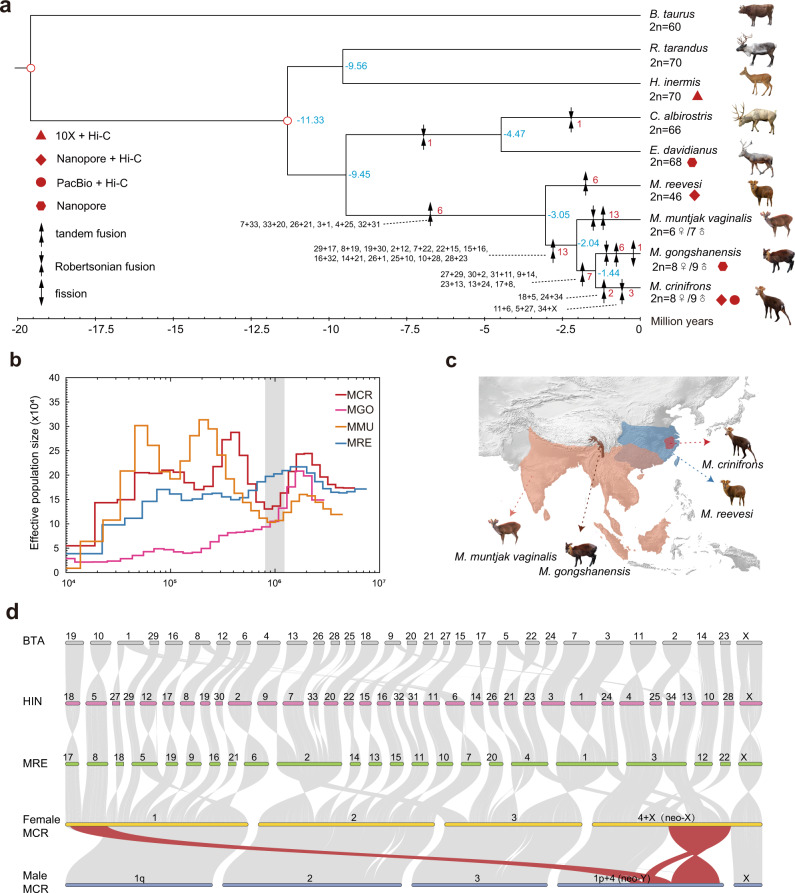
Fig. 1. Phylogeny, demographic histories, and distribution and chromosome synteny of muntjac species.
a Maximum likelihood tree of muntjac and outgroup species with the respective sequencing technologies (red geometries), the divergence time (blue numbers) and number of chromosome fusion or fission events (red numbers) shown. Different combinations of black arrows represent different types of chromosome fusion and fission. The 31 fusion events leading to M. crinifrons are displayed in detail with the chromosome code (black numbers) of H. inermis, which are connected with the arrow mark on the phylogenic tree with dotted lines. Red hollow circles mark the nodes whose divergence times were used as calibration for estimating the divergence time among other species. b The demographic histories of M. reevesi (MRE), M. muntjak vaginalis (MMU), M. gongshanensis (MGO), and M. crinifrons (MCR) estimated by PSMC37. The gray box marks the time range of the Xixiabangma Glaciation (XG, 0.8-1.17 million years ago). c Topographic map on current geographic distribution of the four muntjac species. The colors of dashed line are consistent with the colors of distribution areas of a particular species. d The chromosome synteny between B. taurus, H. inermis, M. reevesi, female and male M. crinifrons with chromosome names shown above. 1p and 1q represent short arm and long arm of chromosome 1, respectively. The red line indicates the synteny blocks of female and male M. crinifrons in neo-Y inverted regions.