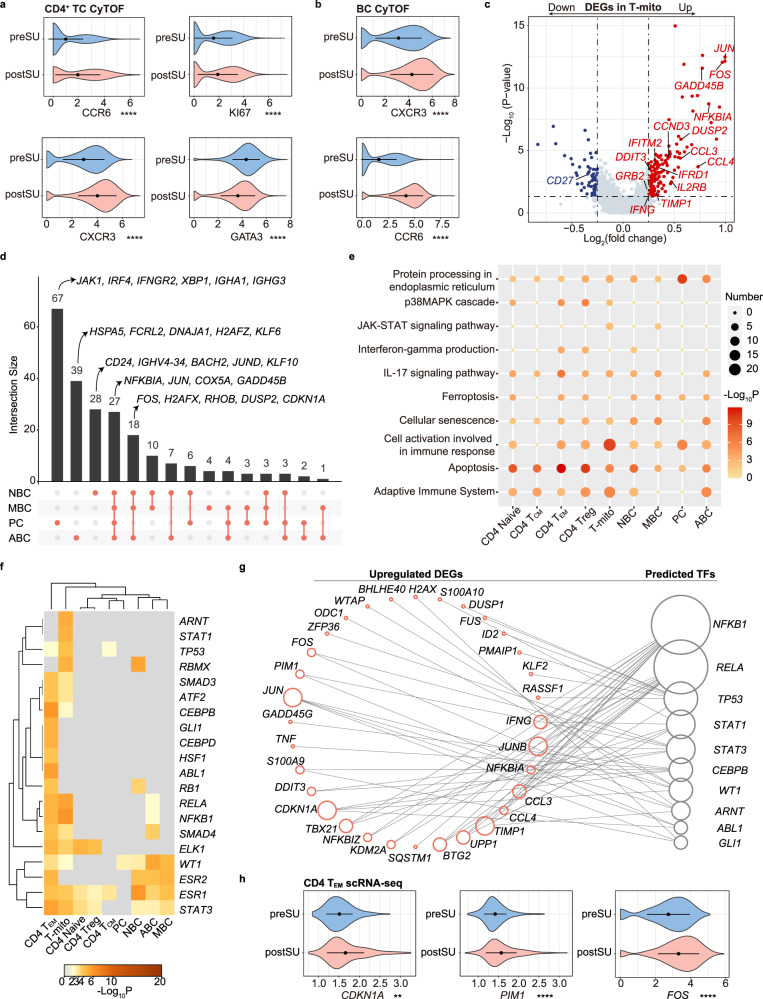
Fig. 3. Changes in proteomic and transcriptional profiles of CD4+ TC, T-mito and BC.
a Violin plot showing the expression of CCR6, CXCR3, KI67, GATA3 in CD4+ TC between preSU and postSU groups in CyTOF. b Violin plot showing the expression of CXCR3, CCR6 in BC between preSU and postSU groups in CyTOF. c Volcano plot showing DEGs of T-mito between preSU and postSU groups. d UpSet Plot showing the integrated comparative analysis of upregulated DEGs in BC subsets. e Representative GO biological process and pathways enriched in upregulated DEGs based on functional enrichment analysis in CD4+ TC, T-mito and BC subsets. f The heatmap showing the enhanced activity of TFs predicted by TRRUST analysis in CD4+ TC, T-mito and BC subsets. g Network visualization of the predicted transcriptional regulatory networks enhanced by SU using TRRUST tool. h Violin plot showing the expression of CDKN1A, PIM1, FOS in CD4 TEM between preSU and postSU groups in scRNA-seq. For the box plot within each violin plot, middle lines indicate median values, boxes range from the 25th to 75th percentiles. Significance in a, b and h was calculated using two-sided Wilcoxon test as implemented in the function “compare_means” with default parameters; **P < 0.01, ****P < 0.0001.