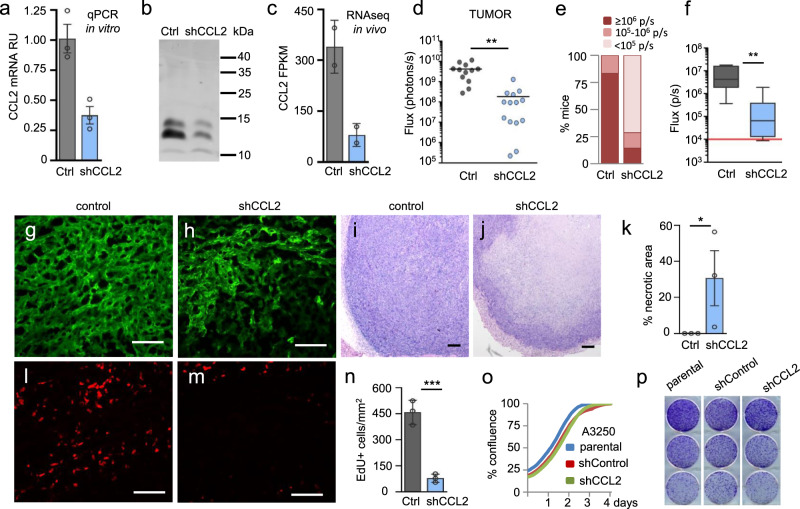
Fig. 8. Effects of CCL2 knockdown on A3250 tumor growth and metastasis.
a CCL2 mRNA expression levels in A3250 control and A3250-shCCL2 cells determined by qPCR. n = 3 biologically independent samples. Data are presented as mean values ± SD. Two independent experiments were performed. b CCL2 Western analysis of A3250 cells in vitro as indicated. c CCL2 mRNA levels in A3250-control and A3250-shCCL2 tumors determined by mRNA-Seq. n = 2 biologically independent samples. Data are presented as mean values ± SD. Two independent experiments were performed. d Bioluminescence imaging (total Flux) of A3250-control (n = 12 tumors from 6 mice) and A3250-shCCL2 tumors (n = 14 tumors from 7 mice) in vivo 6 weeks post-injection. Two tumors were harvested from each mouse, therefore to control for repeated measures, a Linear Mixed Model fit by REML and t test using Satterthwaite’s method was used for analysis (p = 0.00115). e Metastatic burden in the lungs of A3250-control (n = 6) and A3250-shCCL2-bearing (n = 7) tumors determined by ex vivo bioluminescence imaging at 6 weeks. Signal intensity is indicated in photons/second (p/s). Incidence of metastases: macro-metastases, ≥106 p/s; micro-metastases, 105–106 p/s; disseminated tumor cells, 104–105 p/s. f Box and whisker plot showing total metastatic burden by bioluminescence in a mouse cohort as indicated (n = 6 A3250-control and n = 7 A3250-shCCL2 biologically independent lung samples). Solid black lines denote mean values. Red solid line indicates the threshold of positive Fluc signal above background. A two-tailed Mann–Whitney t test yielded p = 0.005. g, h Viable tumor area of control and A3250-shCCL2 tumor sections visualized by GFP. i, j H&E staining showing viable and necrotic tumor areas in tumors as indicated. k Quantification of necrotic tumor areas based on H&E staining (n = 3 biologically independent tumors per cell line). Data are presented as mean values ± SEM. P = 0.0453, determined through a one sample two-tailed t test. l, m EdU labeled proliferating cells (red) in sections of control and shCCL2 tumors shown in (g) and (h). n Proliferation of tumor cells in vivo, determined by EdU quantification on GFP+ tumor areas. n = 3 biologically independent tumors per cell line. Data is presented as mean values ± SD. A two-tailed unpaired t test yielded p = 0.0009. Images are representative of two independent experiments. o Proliferation of tumor cells in vitro. n = 3 biologically independent samples per condition. Data are representative of two independent experiments. p Colony Forming Assay on plastic. n = 3 biologically independent samples per condition. Images are representative of two independent experiments. All scale bars 100 μm. **p < 0.05 **p < 0.01 ***p < 0.001. RU = Relative Units; FPKM = Fragments Per Kilobase of exon per Million mapped reads. Source data are provided as a Source Data file.