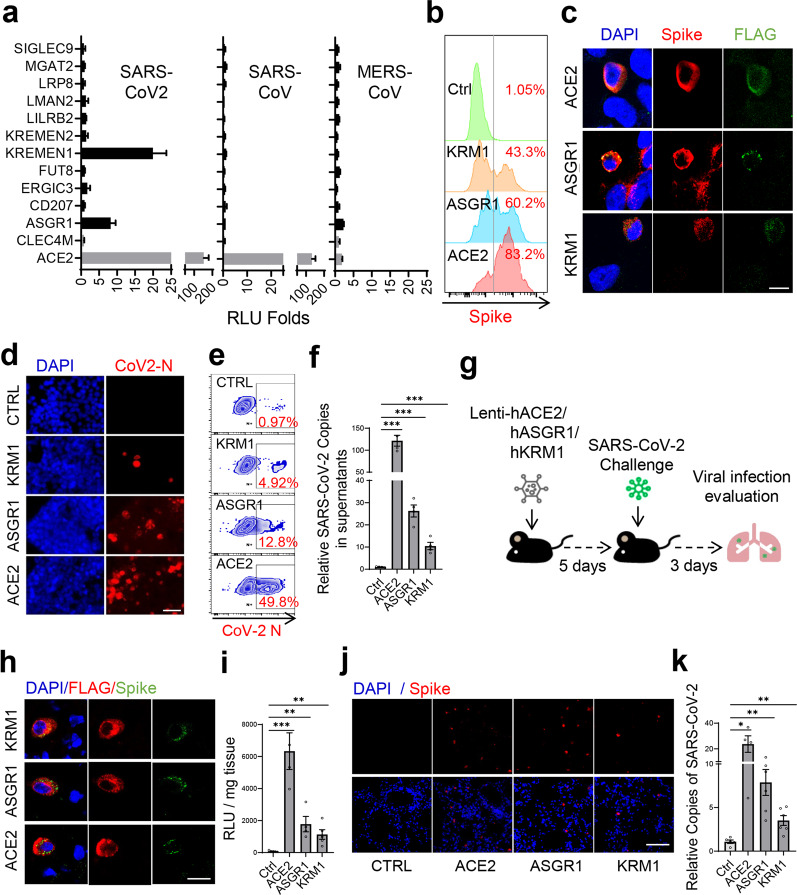
Fig. 2. KREMEN1 and ASGR1 directly mediate SARS-CoV-2 entry both in vitro and in vivo.
a Identified S-binding membrane proteins were individually ectopically expressed in ACE2-KO HEK293T cells, and the cells were then infected with S-pseudotyped SARS-CoV-2, SARS-CoV, and MERS-CoV, separately. Relative luciferase activities (RLU) to that of empty vector-transfected cells were measured 60 h post-infection (means ± SD, n = 3). b KREMEN1-, ASGR1-, or ACE2-transfected ACE2-KO HEK293T cells were incubated with S-pseudotyped SARS-CoV-2 on ice for 1 h, and viral attachment to cell surface was measured by spike staining and flow cytometry. c ACE2-KO HEK293T cells expressing FLAG-tagged KREMEN1, ASGR1, or ACE2 were infected with S-pseudotyped SARS-CoV-2 for 48 h, immunofluorescence was performed with antibodies against FLAG and S protein (scale bar, 50 μm). d–f KREMEN1-, ASGR1-, or ACE2-transfected ACE2-KO HEK293T cells were infected with authentic SARS-CoV-2, and immunofluorescence (d) and flow cytometry (e) were performed with an antibody against the N protein of SARS-CoV-2 72 h post-infection (scale bar, 50 μm); the viral titers in the cell supernatants (f) were measured by RT-qPCR of SARS-CoV-2 ORF1ab gene (means ± SEM, n = 4). g Experimental strategy to study the role of ASK receptors in mediating SARS-CoV-2 infection in mouse models. h, i Immunofluorescence staining of S protein (anti-S) and ASK receptors (anti-FLAG) (h) of the lungs of Lenti-ACE2/ASGR1/KREMEN1-transduced mice challenged with S-pseudotyped SARS-CoV-2 (scale bar, 200 μm); the viral loads in the lungs of each group (i) were measured by luciferase assay (means ± SEM, n = 4–6 biologically independent mice from each group). j, k S protein staining (j) of the lungs of Lenti-ACE2/ASGR1/KREMEN1-transduced mice challenged with SARS-CoV-2 (scale bar, 200 μm); the viral loads in the lungs of each group (k) were measured by RT-qPCR of the SARS-CoV-2 ORF1ab gene (means ± SEM, n = 4–6 biologically independent mice from each group). The statistical significance was evaluated by unpaired two-tailed Student’s t-tests, *P < 0.05, **P < 0.01, ***P < 0.001.