Significance
Down syndrome (DS) neurocognitive disabilities associated with trisomy 21 are known; however, gene changes within individual brain cells occurring with age are unknown. Here, we interrogated >170,000 cells from 29 aging DS and control brains using single-nucleus RNA sequencing. We observed increases in inhibitory-over-excitatory neurons, microglial activation in the youngest DS brains coinciding with overexpression of genes associated with microglial-mediated synaptic pruning, and overexpression of the chromosome 21 gene RUNX1 that may be a potential driving factor in microglial activation. Single-nucleus long-read sequencing revealed hundreds of thousands of unannotated RNA transcripts. These included diverse species for the Alzheimer’s disease gene—amyloid precursor protein—that contained intra-exonic junctions previously associated with somatic gene recombination, which was also identified in ∼8,000 other genes.
Keywords: Down syndrome, single-nucleus RNA sequencing, microglia, intra-exonic junctions, gene recombination
Abstract
Down syndrome (DS), trisomy of human chromosome 21 (HSA21), is characterized by lifelong cognitive impairments and the development of the neuropathological hallmarks of Alzheimer’s disease (AD). The cellular and molecular modifications responsible for these effects are not understood. Here we performed single-nucleus RNA sequencing (snRNA-seq) employing both short- (Illumina) and long-read (Pacific Biosciences) sequencing technologies on a total of 29 DS and non-DS control prefrontal cortex samples. In DS, the ratio of inhibitory-to-excitatory neurons was significantly increased, which was not observed in previous reports examining sporadic AD. DS microglial transcriptomes displayed AD-related aging and activation signatures in advance of AD neuropathology, with increased microglial expression of C1q complement genes (associated with dendritic pruning) and the HSA21 transcription factor gene RUNX1. Long-read sequencing detected vast RNA isoform diversity within and among specific cell types, including numerous sequences that differed between DS and control brains. Notably, over 8,000 genes produced RNAs containing intra-exonic junctions, including amyloid precursor protein (APP) that had previously been associated with somatic gene recombination. These and related results illuminate large-scale cellular and transcriptomic alterations as features of the aging DS brain.
Down syndrome (DS) is a common genetic disorder affecting ∼1 in 700 live births (1). It is caused by the triplication of human chromosome 21 (HSA21) and results in numerous impairments. Brain abnormalities produce deficits in cognitive performance, learning, and language acquisition, as well as short- and long-term memory impairment (2). As DS individuals age, they show increased incidence of dementia and neuropathological hallmarks of Alzheimer’s disease (AD) by their 40s (3). The mechanistic etiology of the complex DS phenotype is known only in part. It includes defects in neuronal development (4) and GABA signaling (5). Alterations in dendritic spine dynamics have also been reported in DS models (6–8). It is hypothesized that the early onset of AD neuropathology and dementia in DS is driven by overexpression of genes located on HSA21, such as the kinase DYRK1A and especially amyloid precursor protein (APP) (9). Notably, increased brain transcription (9) and increased copy numbers of the APP gene have been linked to APP somatic gene recombination associated with sporadic AD. This form of gene recombination produced internally truncated RNA sequences containing intra-exonic junctions (IEJs) (10).
Single-cell sequencing technologies have opened new avenues to understanding cellular transcriptomics, particularly through the use of single-nucleus RNA sequencing (snRNA-seq), which has been applied to normal (11, 12) and diseased (13–15) human brains, but has not been reported for postnatal or aging DS brains. Bulk RNA sequencing (RNA-seq) studies of DS brains identified global alterations in gene expression (16, 17), but how specific cell types or RNA isoforms are impacted remains unknown. In addition, nearly all single-cell or single-nucleus transcriptomic studies lack unbiased RNA isoform information, which may have important biological consequences for cellular function (18, 19). Here, we report single-nucleus analyses using both short- and long-read snRNA-seq on aging DS brains versus non-DS controls. These results revealed significant differences between aging DS and control brains in regard to their cellular composition and isoform-specific transcriptomes, including novel truncated RNAs containing IEJs and involving not only APP but thousands of other genes.
Results
DS and Control Brain Sample Characteristics.
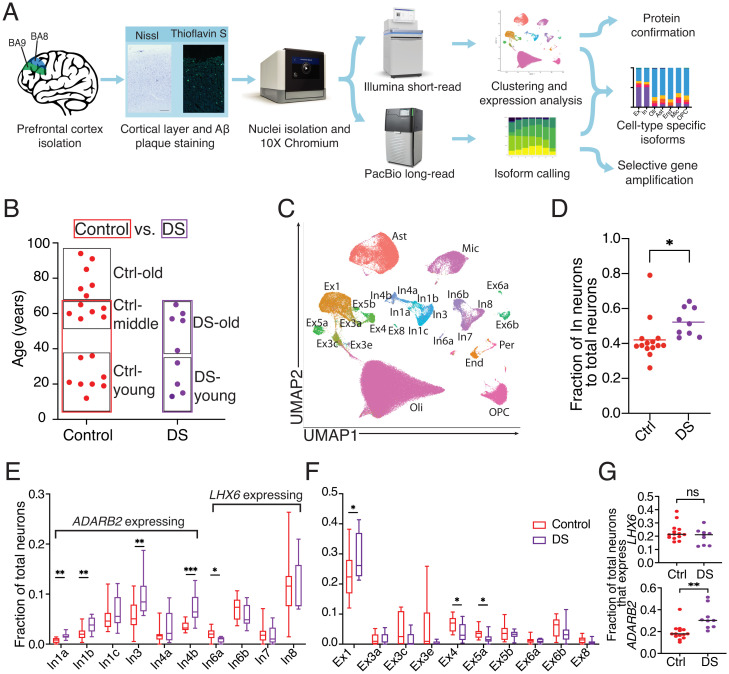
snRNA-seq was performed on DS and control samples (Fig. 1A). Human cerebral cortical Brodmann Areas 8/9 (BAs 8/9) from 56 DS and control brains were sectioned and assessed for cortical layers, RNA integrity number (RIN), and defined neuropathological signs of AD (SI Appendix, Fig. S1A and Dataset S1). The prefrontal cortex was profiled because it is essential to memory and behavior, and gene-expression differences could offer key insights into the DS brain (20). Samples with the confirmed presence of all six cortical layers and RIN ≥ 6 were included in subsequent sequencing experiments. Samples with RIN < 6 were excluded from analysis because of negative trends in key single-cell output characteristics, including the number of captured nuclei, total genes detected, and median genes per nucleus (SI Appendix, Fig. S1B). Twenty-nine samples met the inclusion criteria (Fig. 1B) and were processed for snRNA-seq. Nine DS and 14 control samples were matched for age, sex, and RIN and were used for primary analyses (SI Appendix, Fig. S1C and Dataset S1) (Mann–Whitney U test, P > 0.1). Brains were categorized as “young” if they were ≤36 y of age, and thioflavin-S staining confirmed a lack of neuropathological hallmarks of AD (SI Appendix, Fig. S1D). Another six control brains older than 70 y (“Ctrl-old”) were processed for snRNA-seq to profile aging in control brains. In addition to matching samples for age, sex, and RIN, potential batch effects were accounted for by randomizing samples during processing and utilizing Seurat v3 for analysis (Materials and Methods) (21).
Fig. 1.
Experimental approach for cell clustering and altered neuronal fractions in DS. (A) Experimental outline for selecting samples and processing short- and long-read sequencing of snRNA-seq data. (B) Ages for all samples analyzed by snRNA-seq and sample groups used in subsequent analyses (boxed). (C) UMAP and cell-type assignments of nuclei from DS and control age-matched brains. (D) Fraction of total neurons identified as inhibitory (In) in control and DS brains. (E and F) Fraction of inhibitory (E) and excitatory (Ex) (F) neuronal subtypes in control and DS brains. For E and F, boxes extend from the 25th to 75th percentiles and whiskers extend from minimum to maximum values. (G) Fraction of inhibitory neurons that expressed LHX6 or ADARB2. For D–G, asterisks denote statistical significance in unpaired t test (*P < 0.05, **P < 0.01, ***P < 0.001).
A total of 172,237 filtered transcriptomic cell profiles were generated from snRNA-seq cDNA libraries (using 10X Genomics Single Cell 3′ v3 system) and clustered using Seurat v3 (22). Clusters were identified with marker genes previously established in the human prefrontal cortex and labeled as: astrocytes (Ast), endothelial cells (End), excitatory neurons (Ex1-8), inhibitory neurons (In1-8), microglia (Mic), oligodendrocytes (Oli), oligodendrocyte precursor cells (OPC), and pericytes (Per) (11) (Fig. 1C). This approach labeled 20 known neuronal subclusters, while only 11 were resolved by unbiased techniques (SI Appendix, Fig. S1E). Clusters displayed a gene-expression pattern similar to previous snRNA-seq classifications within the human brain (SI Appendix, Fig. S1F and Dataset S2). All major cell types were present in each cohort and not significantly altered by sex or processing batch (SI Appendix, Fig. S1 G–I and Dataset S1). Prefragmented samples from the same cDNA libraries used for short-read sequencing enabled generation of ∼98 million long reads that revealed 434,201 unique RNA isoforms with cellular barcodes.
Increased Inhibitory:Excitatory Neuron Ratios and Neuronal Subtype Alterations in DS Prefrontal Cortex.
An imbalance in inhibitory vs. excitatory neuronal firing has been reported in mouse models of DS (23). However, it is unclear if such an imbalance exists in human DS. At all examined ages, the proportion of inhibitory to total neurons was significantly increased in DS brains compared with controls (Fig. 1D) (unpaired t test, P = 0.04). A multiple linear regression analysis accounting for sex, RIN, age, and DS vs. control status was also performed. DS was the only variable with a significant effect (P = 0.03). Immunolabeling for inhibitory neurons supported the snRNA-seq data (SI Appendix, Fig. S2 A and B). An inhibitory:excitatory imbalance was not observed in published AD datasets (13) (SI Appendix, Fig. S2C), potentially indicating that this is a DS-specific phenomenon. Notably, this imbalance was previously observed in a single-cell assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) analysis of the Ts65Dn mouse model of DS (24) (SI Appendix, Fig. S2D), further supporting the imbalance as a feature of the DS brain. Focused analyses on DS-young vs. Ctrl-young samples identified similar results (SI Appendix, Fig. S2E), supporting early changes in DS. The proportions of inhibitory neurons from the In1a, In1b, In3, and In4b clusters were increased compared with controls (Fig. 1E). By comparison, proportions of excitatory neuronal clusters were relatively unchanged or slightly reduced at all ages with the exception of the Ex1 subcluster (Fig. 1F) (11).
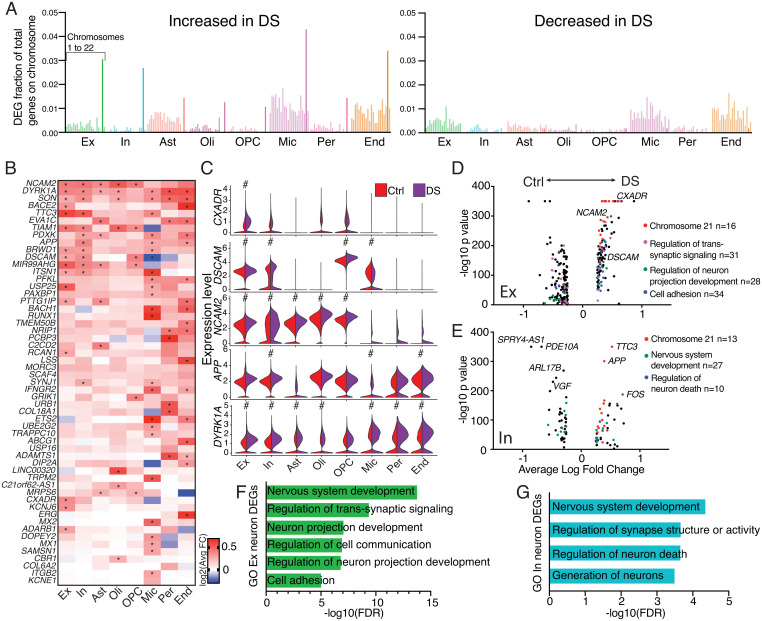
Fig. 2.
Gene-expression changes in DS. (A) Bar graphs displaying the fraction of annotated genes on each chromosome detected as an up-regulated or down-regulated DEG in DS compared with control brains for each specific cell type. Chr21 is bolded. (B) Heatmap displaying the log2 fold-change of select HSA21 genes. Asterisks denote genes that meet the criteria as DEGs (log2 fold-change > 0.25, a Bonferroni adjusted P value < 0.05, and expression in at least 10% of cells within analyzed cell types). (C) Violin plots of five HSA21 genes hypothesized to play central roles in DS; pound (#) symbol denotes genes that meets DEG criteria. (D and E) Volcano plots displaying DEGs for each cell type, color coded by GO biological processes classification. (F and G) Key biological processes determined by GO analyses for excitatory neuron (F) and inhibitory neuron (G) full transcriptome DEGs, FDR defined as false discovery rate P value.
Inhibitory neurons within the cerebral cortex arise developmentally from defined portions of the embryonic ganglionic eminence. In mice, interneurons expressing LHX6 (In6-In8) originate from the medial ganglionic eminence (MGE), while interneurons expressing ADARB2 (In1-In4) are derived from the caudal ganglionic eminence (CGE) (25). A distinction between LHX6- and ADARB2-expressing neurons is also observed in humans (SI Appendix, Fig. S1F). LHX6-expressing neurons were present in the human DS brain at similar proportions to controls, while ADARB2-expressing neurons were overrepresented in DS (Fig. 1G) (unpaired t test, P < 0.01), supporting a CGE origin of neuronal imbalance. These cellular subtype proportional changes were also observed in the DS-young cohort (SI Appendix, Fig. S2 F–H).
Cell-Type–Specific Changes in HSA21 Genes in the DS Brain.
HSA21 trisomy alters expression of HSA21 genes (26). However, direct proportionality between gene copy number and transcription is not expected (16, 17, 27, 28). Comparison of DS with control brains across genes on HSA21 in each individual cell type identified 308 of 4,008 genes with an expression fold-change > 1.1 in DS, and only 9 showed a >1.5-fold increase (SI Appendix, Fig. S3A), signifying limited overexpression of HSA21 genes.
To further investigate HSA21 gene expression in DS brain cell types, data were filtered to focus on differentially expressed genes (DEGs) having a log2 fold-change > 0.25, a Bonferroni adjusted P value < 0.05, and expression in at least 10% of cells within analyzed cell types. A limited number of genes on HSA21 were observed as DEGs, but as expected, significantly more of these DEGs had increased expression in DS (Fig. 2 A and B). Microglia had the greatest number of HSA21 DEGs (DEGs = 25), with endothelial cells (DEGs = 20) and neurons (Ex DEGs = 17; In DEGs = 15) also showing extensive changes in HSA21 gene expression (SI Appendix, Fig. S3A). Differential expression analysis of all genes also identified the greatest expression changes occurring in microglia (SI Appendix, Fig. S3B). Numerous genes on HSA21 were differentially expressed in multiple cell types, including the cell-adhesion molecules, NCAM2 and DSCAM, the splicing regulator, SON, and the kinase, DYRK1A. In neurons, DSCAM (29), CXADR (30), APP (31), and NCAM2 (32) were altered in both excitatory and inhibitory neuronal populations in the DS brain; these genes are directly involved in neuronal cell–cell interactions and neurite outgrowth (Fig. 2C). Notably, DSCAM was specifically expressed in ADARB2-expressing interneurons, potentially playing a key role in the observed overrepresentation of these cells. In contrast, DSCAM showed low expression in LHX6-expressing interneurons (SI Appendix, Fig. S3C), while its expression was significantly down-regulated in DS microglia compared with controls (Fig. 2 B and C).
Dysregulation of Key Neurological Pathways in DS Revealed by Gene Ontology.
To study functional changes in DS neurons, DEGs from the entire transcriptome were analyzed by gene ontology (GO) (33–35) (Fig. 2 D–G). DS excitatory neuron DEGs were involved in the regulation of transsynaptic signaling, the regulation of neuron projection development and cell adhesion (Fig. 2 D and F and SI Appendix, Fig. S3 D and E). Significantly up-regulated genes included the ephrin receptors, EPHA3, EPHA5, and EPHA6, which are involved in neural development (36), the membrane receptors, ROBO1 and ROBO2, and secreted guidance cues, SEMA3C and SLIT2 (Dataset S3), which are all involved in axonal guidance and maintenance of synaptic connections (37).
Inhibitory neuron GO categories included nervous system development and regulation of neuron death (Fig. 2 E and G and SI Appendix, Fig. S2 F and G). Intriguingly, the immediate-early gene FOS, a marker of neuronal firing (38), was the most overexpressed gene not on HSA21 (Fig. 2E), supporting increased inhibitory neuron firing in addition to inhibitory neuron overrepresentation in the DS brain.
Variable Expression of Aging-Associated Genes in Controls.
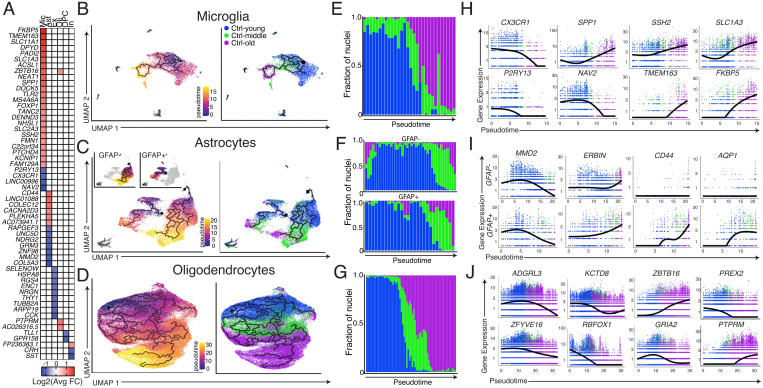
Control brains were used to identify DEGs associated with brain age. No shift in the overall inhibitory:excitatory neuron ratio was observed with age (SI Appendix, Fig. S4A). However, slight changes were observed in specific subtypes of both inhibitory and excitatory neurons, including a decrease in the number of SST-expressing interneurons (In7 and In8) (SI Appendix, Fig. S4 B and C). Aging DEGs were identified from the comparisons between Ctrl-young vs. Ctrl-middle and Ctrl-middle vs. Ctrl-old, and particularly involved microglia and astrocytes (Fig. 3A and Datasets S6–S8). To study the effects of aging on the transcriptome in a continuous manner, an unsupervised pseudotime-trajectory analysis using reversed graph embedding via Monocle 3 (39) was pursued for individual cell types. Microglia, astrocytes, and oligodendrocytes clustered in an age-dependent manner (SI Appendix, Fig. S4D) and displayed pseudotime trajectories that clearly tracked from young to old samples (Fig. 3 B–G). Microglia clustered distinctly by age and showed widespread transcriptomic hallmarks of activation with increasing age (15, 40) (Fig. 3B), including increased expression of the inflammatory mediator, SPP1, and loss of both the chemokine receptor, CX3CR1, and the purinergic receptor, P2RY13 (Fig. 3H).
Fig. 3.
Cell-type-specific signatures of aging in control brains. (A) Heatmap of most differentially expressed aging DEGs for each cell type. (B–D) Unsupervised pseudotime trajectories with cells colored by pseudotime assignment (Left) and age (Right). OPCs are not included in oligodendrocyte analyses. (E–G) Fraction of nuclei that comprises discrete intervals of pseudotime. (H–J) Expression levels of aging DEGs of interest with respect to pseudotime. F includes plots for both the GFAP+ partition and GFAP− partition with respect to pseudotime. In B–G and H–J, each dot represents a single nucleus.
Pseudotime analysis of astrocytes independently partitioned into two trajectories, each proceeding from young to old. One of the groups predominantly expressed two markers of astrocyte activation, GFAP and FOS, and is referred to as GFAP+ (SI Appendix, Fig. S4E). Ctrl-young, Ctrl-middle, and Ctrl-old groups had 26%, 22%, and 39% of total astrocytes partition as GFAP+, respectively, signifying that, unlike microglia, there does not appear to be activation of all astrocytes during aging. However, both GFAP+ and GFAP− partitions showed transcriptomic signs of aging. Genes, including MMD2 and ERBIN, were significantly changed in each trajectory, while others, including the OPN receptor CD44, increased only in GFAP+ astrocytes (Fig. 3I).
As expected, concurrent with exhaustion of the OPC pool, the ratio of OPCs to oligodendrocytes decreased with age (SI Appendix, Fig. S4F). This correlated with decreased expression of the AMPA receptor subunit GRIA2, which has been tied to oligodendrocyte survival and myelination (41) (Fig. 3J).
Early Activation of DS Microglia.
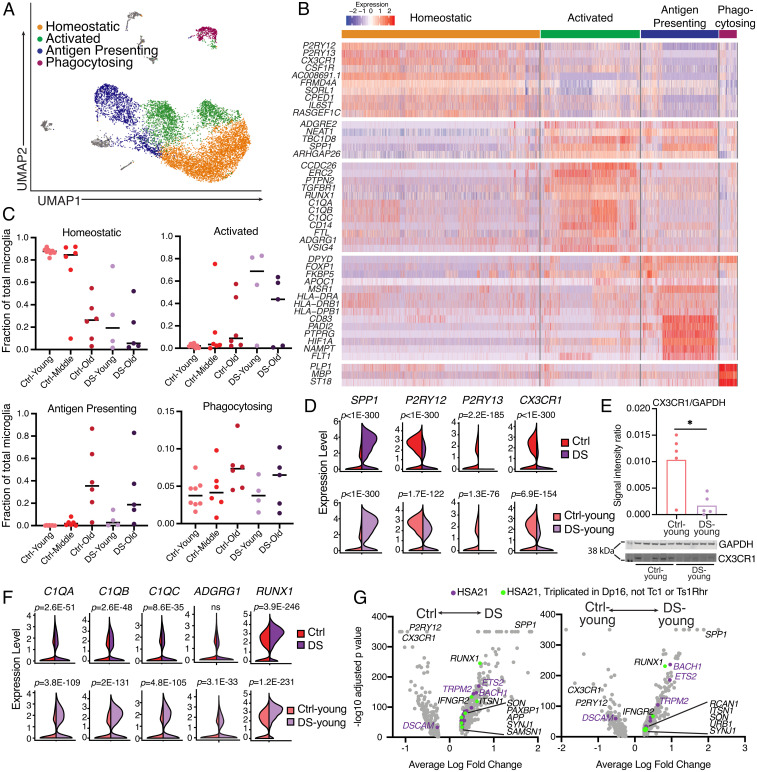
Many DEGs identified in the aging brain were also differentially expressed in DS compared with age-matched control brains, with a striking signal in microglia, where 40 of 45 aging DEGs were also DS DEGs (SI Appendix, Fig. S5A). Pseudotime analysis of microglia from all cohorts also indicated an aged microglial state in DS-young brains (SI Appendix, Fig. S5 B and C). To study microglial gene expression and different activation states, microglia from all samples were clustered separately from other cell types (Fig. 4A). Analyses were focused on clusters containing >2.5% of total microglia, resulting in four distinct microglial clusters with gene-expression profiles similar to other single-nucleus microglial datasets (15, 40) (Fig. 4B). The largest cluster, labeled “Homeostatic,” was comprised of microglia expressing homeostatic markers—including CX3CR1, P2RY12, and P2RY13—while lacking activation markers, such as SPP1. A second cluster, labeled “Activated,” had high expression of complement components, C1QA, C1QB, and C1QC, as well as CD14, ERC2, and PTPN2. The third major cluster, “Antigen presenting,” contained highly expressed genes associated with antigen presentation, including CD83, HLA-DRA, HLA-DRB1, and HLA-DPB1, as well as PADI2, MSR1, and APOC1. Lastly, a small subset of microglia expressed transcripts typically associated with oligodendrocytes, specifically MBP, PLP1, and ST18; these “Phagocytosing” microglia are hypothesized to internalize oligodendrocyte transcripts while phagocytosing myelin (15). Strikingly, > 80% of microglia from every Ctrl-young sample clustered as homeostatic, contrasting with the age-matched DS-young cohort that averaged only 28% (Fig. 4C). DS-young microglia were largely classified as activated (Fig. 4C). As expected, DS-old microglia clustered as both activated and antigen presenting, likely associated with the AD pathology in this cohort (Fig. 4C). Broad increases in expression of microglial activation markers and a loss of homeostatic gene expression were observed in microglia in all DS samples, as well as the DS-young cohort (Fig. 4D). Transcripts of CX3CR1 and C1QA were observed in generally distinct cells (SI Appendix, Fig. S5D), and CX3CR1 transcripts were sequenced primarily in microglia (SI Appendix, Fig. S5E). Significant decreases in CX3CR1 protein were observed in DS-young brains (Fig. 4E).
Fig. 4.
Hallmarks of microglial activation in DS microglia. (A) UMAP of microglia from all processed samples, colored by microglial subcluster. (B) Heatmap displaying key differentially expressed genes used to define microglial subclusters. (C) Fraction of total microglia from each sample that clustered in each of the four major microglia subclusters. (D) Violin plots of gene expression for hallmark microglial activation genes from DS vs. Ctrl and DS-young vs. Ctrl-young cohorts; adjusted P value using Bonferroni correction on Wilcoxon rank sum test. (E) Western blot for CX3CR1 and quantification relative to GAPDH. Asterisk denotes statistical significance in unpaired t test (P = 0.011). (F) Violin plots of gene expression for C1q complement genes, ADGRG1, and RUNX1. (G) Volcano plots for total DEGs in microglia (gray), DEGs from HSA21 that are triplicated in the Dp16 mouse model but not Tc1 or Ts1Rhr (green), and all other HSA21 microglial DEGs (purple) (48–50).
AD-Associated Gene Up-Regulation in DS Microglia.
AD neuropathology uniformly occurs in DS individuals beyond age 40. A direct comparison of DS microglia with human AD microglia (13) revealed a shared increase in APOE and PTPRG expression (SI Appendix, Fig. S5F). However, DS microglia displayed distinct profiles wherein most genes that were down-regulated in AD showed up-regulation in DS (SI Appendix, Fig. S5F), and numerous DEGs in DS microglia were not identified as DEGs in AD. Microglial gene expression was also compared with previously defined disease-associated microglia (DAM) expression signatures (42) (SI Appendix, Fig. S5G). Antigen presenting microglia most closely resembled DAMs, displaying increased expression of many DAM up-regulated DEGs and decreased expression of DAM down-regulated DEGs.
As previously noted, the DS-young cohort lacked the pathological hallmarks of AD. However, microglial genes associated with the earliest signs of AD onset (13) were up-regulated in DS-young microglia, including VSIG4, ADGRG1, CACNA1A, and C1QC (SI Appendix, Fig. S5H), supporting overlap of microglial AD-like activation occurring in the young DS brain. With increasing age, DS microglia showed reductions in complement-associated genes and accompanying increases in antigen presentation-associated genes, like the major histocompatibility complex and CD83 (SI Appendix, Fig. S5I). These results support precocious and evolving microglial activation states with age, modified for differing age-dependent activities, including in response to developing AD.
Increased Expression in DS of Microglial Genes Related to Synaptic Function.
Microglia are implicated in synapse and memory loss through both complement-mediated (43, 44) and ADGRG1-mediated (45) pathways. Remarkably, all three gene components of complement C1q (C1QA, C1QB, C1QC) as well as ADGRG1 were overexpressed in DS microglia, particularly in the DS-young cohort (Fig. 4F and SI Appendix, Fig. S5J), suggesting that overactive pruning by microglia may occur in DS. In addition, a decrease of P2RY12 expression was identified (Fig. 4D); loss or inhibition of P2RY12 has been linked to impaired synaptic function (46). These microglial transcriptomic alterations affecting neurons may contribute to neurocognitive changes in DS.
A significant decrease in the density of dendritic spines was reported in the Dp16 mouse model of DS, which could be reversed by the depletion or inhibition of microglia (47). However, these findings directly conflict with data from the Tc1 and Ts1Rhr mouse models of DS that show no changes in dendritic spine density (7). To discern if an HSA21 gene might be responsible for this discrepancy, we profiled the HSA21 DEGs in human microglia and cross-referenced these with genes triplicated in the Dp16 model, but functionally diploid in Tc1 and Ts1Rhr mice (48–50). Multiple microglial DEGs were triplicated in Dp16 but not Tc1 or Ts1Rhr mice. These genes included the interferon (IFN) receptor, IFNGR2, the splicing regulator, SON, and most significantly, the transcription factor, RUNX1 (Fig. 4G). RUNX1 overexpression was observed broadly in microglia across all DS samples (SI Appendix, Fig. S5J) and was the most overexpressed HSA21 microglial DEG (Fig. 4G). RUNX1 is a key transcription factor in regulating microglial gene expression (51), and its expression typically decreases after early neurodevelopment but can be induced following brain injury in adults (52). TRPM2, a calcium channel that has been tied to microglia activation (53), was one of the top microglial DEGs but is not triplicated in any of the three mouse models discussed, signifying that microglial activation may be even more striking in the human brain. Furthermore, BACH1, triplicated in Tc1 and Dp16 mice, encodes a transcriptional repressor involved in the development of numerous antigen-presenting cell subtypes including macrophages, and its diminished expression correlates with protective autoimmune effects (54, 55), which may be relevant to microglial activation states.
Transcriptomic Diversity Discovered Through Long-Read Sequencing of Single-Nucleus cDNA Libraries.
Long-read single molecular real-time (SMRT) sequencing enables profiling of full-length RNA isoforms from single-cell cDNA libraries (56, 57). Single-nucleus cDNA libraries were sequenced with SMRT sequencing to obtain ∼98 million long reads from 16 individual brains, 8 each from control and DS cohorts (SI Appendix, Fig. S6A). Each sample was sequenced to a depth of 5.5 to 7.5 million raw long reads. Of these, 34,988,576 total reads had both a cellular barcode and a unique molecular identifier (UMI). Reads were analyzed using cDNA_Cupcake and SQANTI2 (58) to identify isoforms and group them into four main categories defined as: full splice match (FSM) isoforms that match GENCODE v28 annotations; incomplete splice match (ISM) isoforms that only partially match annotations and result from 3′ and 5′ truncations; novel in catalog (NIC) isoforms that have not been annotated but contain known splice sites and exons; and novel not in catalog (NNC) isoforms that contain at least one novel splice site. After filtering, 434,201 unique isoforms remained, supported by a total of 6,905,832 reads (SI Appendix, Fig. S6B), and 47.7% of these isoforms were supported by at least two reads with distinct UMIs. Matching the cell barcodes back to the originating cell identified by short-read sequencing enabled cell-type identification for 40.42% of the isoform reads. A majority of the reads that were not associated with a cell type had a barcode that either corresponded to a cell that did not pass quality control in the Seurat analysis (64.7% of unidentified reads) or was determined to be background in the cellranger analysis (25.4%). A small percentage of these reads contained known 10X Genomics cellular barcodes that were not observed in the short-read dataset (1.6%). The remaining reads had cellular barcodes that were not among the available 10X Genomics cellular barcodes and could have resulted from error introduced during library preparation or sequencing (8.3%). Interestingly, long-read coverage was enough to identify most cell types (SI Appendix, Supplementary Text).
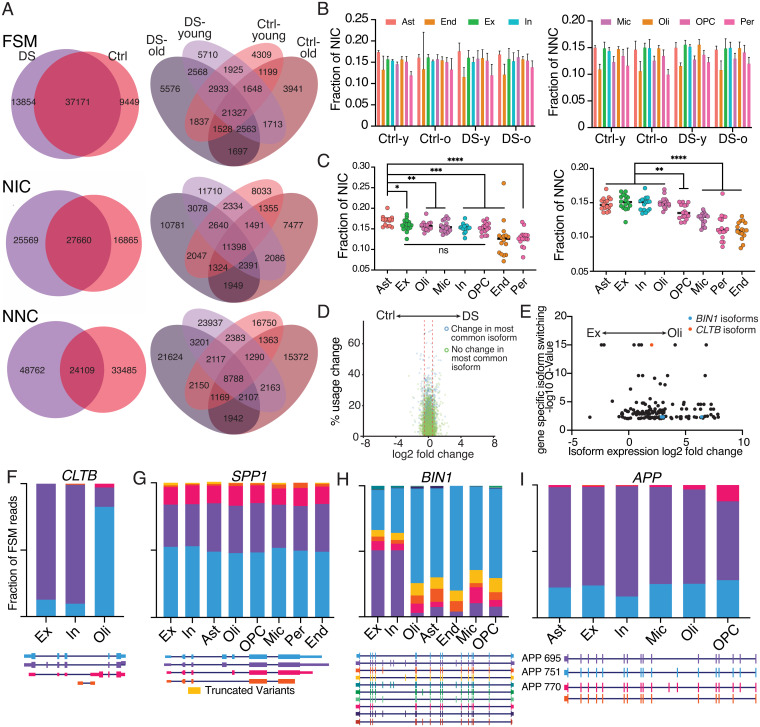
Vast isoform diversity was observed in the brain. Novel isoforms (NIC and NNC) displayed greater variation than annotated forms (FSM) (Fig. 5A), but the overall proportion of novel isoforms did not change with age or across DS and control cohorts (Fig. 5B) (two-way ANOVA, NIC by cohort P = 0.96, NNC by cohort P = 0.13). However, the proportion of novel isoforms did vary with cell type. Analysis of all cells showed the greatest NIC and NNC isoform diversity in astrocytes, whereas endothelial cells and pericytes showed the least (Fig. 5C) (NIC by cell type P < 0.0001, NNC by cell type P < 0.0001). Excitatory neurons, inhibitory neurons, and oligodendrocytes also showed NNC enrichment (Fig. 5C). Multiple types of NNC isoforms were observed and included features such as: novel exon junctions within an intron that created an entirely new exon that does not overlap with any previously annotated exon sequences; IEJs (10) that were formed by joining the internal regions of two exons; and intron retention junctions that were formed by a new splice site that extends exon coordinates partially into the next intron (SI Appendix, Fig. S6 C–E) [IEJs were found in over 8,000 genes (Dataset S10)]. Notably, prior studies using cap analysis gene expression sequencing demonstrated that 94% of NIC and 87% of NNC reads have 5′ ends that contain the transcription start site (59), supporting the conclusion that NIC and NNC reads are indeed bona fide transcripts and not sequencing artifacts.
Fig. 5.
Novel isoform variants and specific isoform changes in different brain cell types. (A) Overlap of FSM, NIC, and NNC isoforms across DS, aging, and control (Ctrl) cohorts. (B) Fraction of total isoforms called as NIC or NNC by cohort and cell type. Two-way ANOVA, NIC by cohort P = 0.96, NIC by cell type P < 0.0001, NNC by cohort P = 0.13, NNC by cell type P < 0.0001. Error bars represent one SD. (C) Fraction of isoforms classified as NIC or NNC for each sample by cell type. Asterisks denote statistical significance in an unpaired t test (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (D) Volcano plot of isoform usage differences between DS and control cohorts. Percent usage change represents the redistribution of isoform proportions within a gene. Log2 fold-change is the change in expression of an isoform between two conditions. (E) Plot of significantly changed isoforms between excitatory and oligodendrocyte populations. Q-value represents the false-discovery rate for which at least one isoform of a particular gene displays differential proportionality across compared groups, signifying changes in isoform usage at the gene level. Only isoforms with <0.01 Q value and nonzero expression in both cell types are displayed. (F–I) Cell-type-specific isoform proportions for cell types with over 50 unique reads mapping to FSM isoforms for CLTB (F), SPP1 (G), BIN1 (H), and APP (I).
Differential Isoform Expression and Usage Are Observed Across Cell Types.
Identification of specific isoforms that are differentially expressed could potentially offer targets for modern therapeutics, such as antisense oligonucleotides or gene therapies (60, 61). Differential isoform expression and proportional isoform usage were analyzed using tappAS (62). Limited differential isoform expression or differential isoform usage was identified between control and DS samples (Fig. 5D). However, pairwise comparisons of isoform usage between cell types revealed numerous genes for which a cell type preferentially utilized one isoform over others (Fig. 5E and SI Appendix, Fig. S7A). Numerous genes switched cell-type–specific isoforms—including SEPT8, RPL13 (SI Appendix, Fig. S7 B and C), and CLTB (Fig. 5F)—which plays an important role in clathrin-mediated endocytosis and utilized a different isoform in neurons compared with oligodendrocytes.
To characterize isoform diversity further, SPP1, BIN1, and APP were selectively amplified from single-nucleus cDNA libraries using the Read1 primer from the 10X adapter and primers designed against their 5′UTRs. Minimal changes in the proportional expression of SPP1 FSM isoforms across cell types (Fig. 5G) or between DS and control cohorts were detected (SI Appendix, Fig. S7D). A significantly higher proportion of reads were found originating from microglia in DS as compared with controls (SI Appendix, Fig. S7E), which is consistent with the differential expression displayed in the short-read data. Untargeted sequencing identified a SPP1 NNC isoform with a novel exon that was confirmed with targeted sequencing. Targeted sequencing also identified an additional four isoforms containing this exon; altogether, this novel exon was supported by 73 UMIs (SI Appendix, Fig. S7F).
In genome-wide association studies, mutations in a region upstream of BIN1 showed the second highest odds-ratio for sporadic AD (63, 64). BIN1 transcripts were selectively amplified, which revealed cell-type–specific isoform switching similar to patterns reported in mice (56). The shortest isoform that lacks all alternatively spliced exons was predominantly sequenced in nonneuronal cell types, whereas the longest isoform that contains all alternatively spliced exons was the predominant isoform in both excitatory and inhibitory neurons (Fig. 5H).
The AD-associated gene, APP, expresses two major brain isoforms, encoding APP-695 and APP-751, with the literature supporting neuron-specific expression of APP-695 that lacks a Kunitz-type serine protease inhibitory domain, implicating neurons as the source for soluble Aβ in the brain (65, 66). However, APP-695 was observed to be the predominant RNA isoform in all cell types (Fig. 5I). Furthermore, total expression levels of APP are similar across neurons, oligodendrocytes, OPCs, pericytes, and endothelial cells (Fig. 2C), signifying that many cell types in the human brain contribute significantly to the RNA expression of APP-695 rather than just neurons. Isoform diversity also included NNC species containing IEJs in APP, consistent with the literature (10) (SI Appendix, Fig. S8).
Discussion
Transcriptomic effects of HSA21 trisomy at the level of single cells in the postnatal and aging DS brain have not been previously reported. snRNA-seq using short- and long-read sequencing, as well as targeted-gene approaches, revealed differences involving multiple transcriptomic pathways and cell types. Most transcriptomic changes affected non-HSA21 genes, supporting global effects of HSA21 trisomy on the transcriptome. However, notable exceptions included APP, NCAM2, DYRK1A, SON, BACE2, and TTC3, indicating dosage effects on select HSA21 genes within specific cell types. Increased neuronal inhibitory:excitatory ratios and increased neurodevelopmental gene expression existed at all examined ages in DS. Prominently, microglia exhibited transcriptomic states indicative of activation. Cell-autonomous causes could include overexpression of the HSA21 transcription factors RUNX1 and BACH1. Furthermore, increased C1q expression could directly affect neuronal process pruning. Aging signatures in the DS brain paralleled gene-expression patterns reported in multiple neurological disorders, particularly AD (13, 42, 67, 68).
The present study represents an extensive single-nucleus transcriptome analysis of the postnatal human DS brain and the largest single-nucleus profiling of RNA isoforms in the human brain to date. Limitations of this study include focused analyses of BA8,9 of the prefrontal cortex, relatively limited numbers of DS brains, and assessment of ∼6,000 cells per brain that while standard, represents a small percentage of total brain cells. These results identified trends that will benefit from expanded analyses in the future. At least three identified features deserve additional comment.
First, neurodevelopmental transcriptome differences are prominent in DS brain cells, including those of cell adhesion genes like DSCAM (29), CXADR (30), APP (31), and NCAM2 (32), as well as genes of the Robo-Slit-Ephrin pathways that normally contribute to axonal guidance, synapse formation, and neurogenesis (36, 37). Abnormal neurodevelopmental programs in DS are further supported by increased ADARB2-expressing (CGE-derived), but not LHX6-expressing (MGE-derived) inhibitory neuron ratios. CGE-derived neurons migrate prenatally from the CGE to the cortex (69, 70), and this increase could result in enhanced neuronal inhibition as supported by DS animal models (71, 72). In human DS, an imbalance of inhibition and excitation may exist considering the clinical reports of elevated seizure activity (73). This dichotomy may be explained by variations in neuronal subsets, such as the statistically significant increase in one cluster of excitatory neurons, Ex1, contrasting with other Ex clusters in DS, as well as by other possible changes affecting epileptogenic parts of the brain that were not assessed.
Second, microglia show major transcriptomic differences at all ages in DS, indicative of activation even at the youngest age examined. Activation could again reflect cell-autonomous mechanisms potentially resulting from increased RUNX1 expression. Alternatively, microglial activation could be indicative of noncell-autonomous mechanisms activated by mismatched neurodevelopmental activities involving C1q complement and other pruning genes associated with exuberant axon outgrowth and synapse formation/elimination, whereby microglia would face a chronic activating-milieu to remove surplus or mismatched process outgrowth and neuronal connections. The down-regulation of genes like P2RY12 in microglia may also contribute to the increased prevalence of seizures in DS (46). In later adult life, microglial activation could additionally reflect stimuli associated with incipient AD and contribute further to neurocognitive deficits. snRNA-seq was sufficient to distinguish activation states, supporting the possibility of a distinct microglial transcriptomic profile compared with AD. The combination of persistent neurodevelopmental gene expression and induced microglial activation provide insight into functional deficits within the DS brain that may be distinguishable from AD signatures.
Third, isoform resolution in single-cell transcriptomic profiling is essential to generating a full understanding of transcriptional biology yet cannot be achieved by standard 3′ short-read sequencing techniques. RNA splice variants have important roles in development and disease (19, 74), and these new data provide an initial platform for approaching cell-type–specific isoforms in the DS and normal brain. Additionally, this study explored the potential for identifying cell types using only long reads, an approach that would eliminate the need for short-read sequencing in cell-type–specific isoform profiling, and suggests that this can be achieved with reasonable accuracy, but would require greater sequencing depth for optimal identification. Notably, isoform diversity and usage varied extensively across cell types, while being relatively stable between disease cohorts, supporting isoform functions in maintaining cell identity. The thousands of novel sequences beyond known splice variants, whose functions are unknown, provide a new reservoir of transcripts toward understanding the normal and diseased brain. These include isoforms with novel structures like IEJs that were detected on APP and over 8,000 thousand additional genes, which might reflect the widespread operation of somatic gene recombination mechanisms, including those relevant to AD (10). Additional experiments are required to determine if these IEJs reflect expression of somatically recombined genes and/or if they are novel splicing variants, which are not mutually exclusive possibilities. Overall, these snRNA-seq studies of the normal aging and DS brain implicate both intrinsic neurodevelopmental cellular processes and RNA isoform diversity, towards providing understanding and identifying therapeutic targets to aid DS individuals.
Materials and Methods
See SI Appendix for more details.
Tissue Sampling and Preparation.
Frozen tissue samples from BA 8 or 9 of the prefrontal cortex were obtained from multiple sources and stored at −80 °C. Samples were sectioned in a cryostat set at −20 °C.
RNA Integrity Measurement.
RNA was isolated using a RNeasy isolation kit from Qiagen and evaluated on an Agilent 4200 TapeStation.
Thioflavin S Staining.
Tissue sections (20 μm) were stained using thioflavin S to visualize amyloid plaques and tau tangles as hallmarks of AD pathology.
Nissl Staining.
Tissue sections (20 μm) were stained using Cresyl violet to visualize the cortical layers of each section.
Nuclei Isolation and Generation of Amplified cDNA Libraries.
DS and control samples were randomized and processed in groups of four to negate potential batch process variation. Tissue sections (300 μm) were removed from frozen storage and immediately submerged in 1 mL of nuclei isolation buffer (20 mM Tris, 320 mM sucrose, 5 mM CaCl2, 3 mM MgAc2, 0.1 mM EDTA, 0.1% Triton-X 100, 0.2% RNase inhibitor) (10, 11). Extracted nuclei were washed twice in PBS + 0.25 mM EGTA + 1% BSA + 0.2% RNase inhibitors (Takara Bio). They were then suspended in PBSE + BSA + RNase inhibitors + 1.25 ug/mL DAPI (Sigma). FANS was performed on a FACSAria Fusion (BD Biosciences) gating out debris from forward scatter and side scatter plots and selecting DAPI+ singlets. Samples were kept on ice until sorting was complete and were immediately processed after sorting. Sorted nuclei were diluted to ∼700 to 1,500 nuclei/mL, and a final concentration was determined using a fluorescent cell counter. The 10X Genomics Single Cell 3′ v3 kit was then used to prepare samples targeting 10,000 single nuclei GEMs (gel bead-in emulsion). The protocol was followed without deviation prior to fragmentation of the cDNA libraries.
cDNA Preparation and Long-Read Sequencing.
Fifty percent of the prefragmented cDNA library was used for long-read sequencing. If the cDNA input concentration was too low for Pacific Biosciences (PacBio) library preparation, the cDNA library was reamplified (Dataset S1) using the same reagents and concentrations as outlined in the 10X Genomics kit protocol. Next, 100 ng of cDNA was used in the PacBio procedure for sequencing. Each sample was sequenced in an individual SMRTcell. An average of 6.003 million polymerase reads were obtained per sample.
Selective Amplification of cDNA Libraries and Subsequent Long-Read Sequencing.
Selective amplification of the genes APP, SPP1, and BIN1 was pursued using custom designed primers and the Read 1 primer from the 10X Genomics preparation. The same cDNA libraries used for long-read analysis were linearly amplified with only the 5′UTR primer present prior to addition of the Read 1 primer. Samples were cleaned with Pronex beads and were sequenced with PacBio Sequel II, as outlined above.
Short-Read snRNA-Seq Data Processing and Filtering.
The 10X Genomics CellRanger software (v3.0.2) was used to demultiplex samples, align reads, quantify UMIs, and generate cell count matrices. Default parameters were used, with the exception of a pre-mRNA reference file (ENSEMBL GRCh38) to capture intronic reads originating from pre-mRNA species present in the nuclei. Using Seurat (v3.0.3), sample matrices were filtered and normalized by the default global-scaling method in Seurat.
Clustering and UMAP Visualization.
Lake et al.’s (11) dataset was used as a reference with Seurat’s TransferData function to label cell types in our samples. Seurat objects from the samples in the same disease/age group were merged (Seurat merge function). For comparisons between two groups, differential expression analysis and pseudotime analysis, merged samples within a group were integrated (Seurat IntegrateData function). The integrated data were then scaled and UMAP embeddings were generated.
GAD67/NeuN Staining.
Tissue sections (20 μm) from DS-young and age matched Ctrl-young samples were costained for GAD67 and NeuN and imaged. After imaging, cells were counted in three separate rows for each section spanning from white matter to the pial surface.
Multiple Linear Regression of Inhibitory:Excitatory Ratios.
For DS and control cohorts, data on sex, RIN, age, and DS vs. control status were collected. Data were input into tables and Prism was utilized to calculate a multiple linear least-squares regression for which the independent variable was ex:in ratio. Sex and DS status were assigned binary indicator variables. No weighting was utilized and no two-way or three-way interactions were accounted for.
DEG Analysis.
Seurat was used to identify DEGs in DS compared with control samples by cell type and between age groups. Default parameters for FindMarkers were used to identify DEGs that were expressed in at least 10% of either of the populations being compared, had at least a 0.25 log fold-difference, and were significant based on a Wilcoxon rank sum test.
Gene Enrichment Analysis.
Pseudotime Analysis.
Count matrices and UMAP projections of specific cell types from Seurat analysis were loaded into Monocle3 (v0.2.1). Cells were partitioned, and pseudotime trajectories were learned and plotted. Endpoints that clustered with the youngest samples’ cells were chosen as the roots for each graph. Differential expression analysis was completed to determine which genes had expression that varied as a function of pseudotime. Astrocytes separated into two partitions that were analyzed individually.
Processing of Long Reads and Isoform Calling.
Samples from both untargeted and targeted long-read datasets were demultiplexed and barcodes were removed using lima (v1.10.0). Following the recommendations in the cDNA_Cupcake repository (version updated 02/07/2020) for single-cell isoform analysis, CCS reads were generated using ccs (v4.2.0) with the following parameters: --minPasses 1 --min-rq 0.8 --minLength 50 --maxLength 21000. The 10X Genomics R1 and TSO primer sequences and reads with improper primer orientation were removed using lima with the parameter --isoseq. UMIs and cellular barcodes were identified for each read. Isoseq3 refine (v3.2.2) was used to remove poly-A tails and artificial concatemers before mapping to the human reference genome (GRCh38). cDNA_Cupcake’s (v9.0.1) collapse_isoforms_by_sam.py was used to collapse redundant isoforms. SQANTI2 (v7.3.2) was used to filter out monoexon isoforms and artifacts of intrapriming and annotate the identified isoforms. Scripts from cDNA_Cupcake were used to assign UMIs/barcodes and isoforms back to specific reads. Original scripts were written to match specific reads back to sample and cell type, summarize which samples each isoform was detected in, and visualize the resulting isoforms in University of California, Santa Cruz (UCSC) Genome Browser.
Differential Isoform Expression and Usage Analysis.
The protocol for using tappAS (v0.99.15) for “Data from Long-read Sequencing Technology” was followed.
RNAscope for Microglial Gene Markers.
Sections of tissue (20 μm) were cut and processed using the recommended kit protocol (2.5 HD Duplex Assay, Advanced Cell Diagnostics 322500). Probes applied were C1QA (485451-C2) and CX3CR1 (411251). Slides were imaged at 40× magnification.
Western Blot Analysis.
Sections (200 μm) of five control and five DS brains (including one with a RIN below the cutoff for sequencing analysis) were lysed in RIPA buffer separated on an Invitrogen Bolt 4–12% Bis-Tris protein gel and transferred to a PVDF membrane. The blot was probed with antibodies to CX3CR1 (Invitrogen #14-6093-81) and GAPDH (Invitrogen #AM4300) and visualized using a LI-COR Biosciences CLx Imager. Bands were quantitated using the LI-COR Image Studio Lite software.
Supplementary Material
Acknowledgments
We thank Richard R. Rivera, Danielle Jones, and Gwendolyn E. Kaeser for their discussions and key input; Laura Wolszon for her efforts to source and obtain human specimens; and Brian James and Kang Liu at the Sanford Burnham Prebys Medical Discovery Institute Genomics Core for RNA integrity number analysis of brain samples. Brain specimens were obtained from the University of Maryland Brain and Tissue Bank, the Goizueta Alzheimer’s Disease Research Center at Emory University, the London Neurodegenerative Diseases Brain Bank, the Newcastle Brain Tissue Resource, and the Southwest Dementia Brain Bank for human brain specimens. We also thank the donors and families who shared these precious brain materials. Research reported in this publication was supported by the National Institute on Aging of the NIH under Awards R56AG073965, R01AG065541, and R01AG071465 (to J.C.) and by the National Institute of General Medical Sciences through the University of California, San Diego Graduate Training Program in Cellular and Molecular Pharmacology institutional training Grant T32 GM007752 (to C.S.L.). This work was also supported by non-federal funds from The Shaffer Family Foundation and The Bruce Ford & Anne Smith Bundy Foundation (to J.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114326118/-/DCSupplemental.
Data Availability
Fastq files for Illumina reads and bam files for PacBio reads are available through the European Genome-Phenome Archive with accession number EGAS00001005691. Isoforms identified by long-read sequencing are available through the UCSC Genome browser:
Untargeted dataset: https://genome.ucsc.edu/s/csl022/DSND_snIsoSeq_sample.
SPP1 targeted dataset: https://genome.ucsc.edu/s/csl022/SPP1_scIsoSeq.
APP targeted dataset: https://genome.ucsc.edu/s/csl022/APP_scIsoSeq.
BIN1 targeted dataset: https://genome.ucsc.edu/s/csl022/BIN1_snIsoSeq.
References
- 1.Parker S. E., et al. ; National Birth Defects Prevention Network, Updated national birth prevalence estimates for selected birth defects in the United States, 2004–2006. Birth Defects Res. A Clin. Mol. Teratol. 88, 1008–1016 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Godfrey M., Lee N. R., Memory profiles in Down syndrome across development: A review of memory abilities through the lifespan. J. Neurodev. Disord. 10, 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wisniewski K. E., Wisniewski H. M., Wen G. Y., Occurrence of neuropathological changes and dementia of Alzheimer’s disease in Down’s syndrome. Ann. Neurol. 17, 278–282 (1985). [DOI] [PubMed] [Google Scholar]
- 4.Haydar T. F., Reeves R. H., Trisomy 21 and early brain development. Trends Neurosci. 35, 81–91 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Contestabile A., Magara S., Cancedda L., The GABAergic hypothesis for cognitive disabilities in Down syndrome. Front. Cell. Neurosci. 11, 54 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiotto A. M. A., et al. , Neuronal cell-intrinsic defects in mouse models of Down syndrome. Front. Neurosci. 13, 1081 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haas M. A., et al. , Alterations to dendritic spine morphology, but not dendrite patterning, of cortical projection neurons in Tc1 and Ts1Rhr mouse models of Down syndrome. PLoS One 8, e78561 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Real R., et al. , In vivo modeling of human neuron dynamics and Down syndrome. Science 362, eaau1810 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiseman F. K., et al. , A genetic cause of Alzheimer disease: Mechanistic insights from Down syndrome. Nat. Rev. Neurosci. 16, 564–574 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee M. H., et al. , Somatic APP gene recombination in Alzheimer’s disease and normal neurons. Nature 563, 639–645 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lake B. B., et al. , Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol. 36, 70–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodge R. D., et al. , Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathys H., et al. , Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velmeshev D., et al. , Single-cell genomics identifies cell type-specific molecular changes in autism. Science 364, 685–689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schirmer L., et al. , Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature 573, 75–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olmos-Serrano J. L., et al. , Down syndrome developmental brain transcriptome reveals defective oligodendrocyte differentiation and myelination. Neuron 89, 1208–1222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lockstone H. E., et al. , Gene expression profiling in the adult Down syndrome brain. Genomics 90, 647–660 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Su C. H., Dhananjaya D., Tarn W. Y., Alternative splicing in neurogenesis and brain development. Front. Mol. Biosci. 5, 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raj B., Blencowe B. J., Alternative splicing in the mammalian nervous system: Recent insights into mechanisms and functional roles. Neuron 87, 14–27 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Fuster J. M., The prefrontal cortex—An update: Time is of the essence. Neuron 30, 319–333 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Tran H. T. N., et al. , A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stuart T., et al. , Comprehensive integration of single-cell data. Cell 177, 1888–1902.e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dierssen M., Down syndrome: The brain in trisomic mode. Nat. Rev. Neurosci. 13, 844–858 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Spektor R., Yang J. W., Lee S., Soloway P. D., Single cell ATAC-seq identifies broad changes in neuronal abundance and chromatin accessibility in Down Syndrome. bioRxiv [Preprint] (2019). https://www.biorxiv.org/content/10.1101/561191v1 (Accessed 15 March 2021). [Google Scholar]
- 25.Tasic B., et al. , Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lana-Elola E., Watson-Scales S. D., Fisher E. M., Tybulewicz V. L., Down syndrome: Searching for the genetic culprits. Dis. Model. Mech. 4, 586–595 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stamoulis G., et al. , Single cell transcriptome in aneuploidies reveals mechanisms of gene dosage imbalance. Nat. Commun. 10, 4495 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veitia R. A., Bottani S., Birchler J. A., Gene dosage effects: Nonlinearities, genetic interactions, and dosage compensation. Trends Genet. 29, 385–393 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Simmons A. B., et al. , DSCAM-mediated control of dendritic and axonal arbor outgrowth enforces tiling and inhibits synaptic plasticity. Proc. Natl. Acad. Sci. U.S.A. 114, E10224–E10233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patzke C., et al. , The coxsackievirus-adenovirus receptor reveals complex homophilic and heterophilic interactions on neural cells. J. Neurosci. 30, 2897–2910 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoe H. S., et al. , Interaction of reelin with amyloid precursor protein promotes neurite outgrowth. J. Neurosci. 29, 7459–7473 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheng L., Leshchyns’ka I., Sytnyk V., Neural cell adhesion molecule 2 promotes the formation of filopodia and neurite branching by inducing submembrane increases in Ca2+ levels. J. Neurosci. 35, 1739–1752 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Gene Ontology Consortium, The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashburner M., et al. ; The Gene Ontology Consortium, Gene ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P. D., PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkinson D. G., Multiple roles of EPH receptors and ephrins in neural development. Nat. Rev. Neurosci. 2, 155–164 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Bashaw G. J., Klein R., Signaling from axon guidance receptors. Cold Spring Harb. Perspect. Biol. 2, a001941 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morgan J. I., Cohen D. R., Hempstead J. L., Curran T., Mapping patterns of c-fos expression in the central nervous system after seizure. Science 237, 192–197 (1987). [DOI] [PubMed] [Google Scholar]
- 39.Qiu X., et al. , Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masuda T., et al. , Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Kougioumtzidou E., et al. , Signalling through AMPA receptors on oligodendrocyte precursors promotes myelination by enhancing oligodendrocyte survival. eLife 6, e28080 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keren-Shaul H., et al. , A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e17 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Stevens B., et al. , The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Wang C., et al. , Microglia mediate forgetting via complement-dependent synaptic elimination. Science 367, 688–694 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Li T., et al. , A splicing isoform of GPR56 mediates microglial synaptic refinement via phosphatidylserine binding. bioRxiv [Preprint] (2020). https://www.biorxiv.org/content/10.1101/2020.04.24.059840v (Accessed 13 March 2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Badimon A., et al. , Negative feedback control of neuronal activity by microglia. Nature 586, 417–423 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinto B., et al. , Rescuing over-activated microglia restores cognitive performance in juvenile animals of the Dp(16) mouse model of Down syndrome. Neuron 108, 887–904.e12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gribble S. M., et al. , Massively parallel sequencing reveals the complex structure of an irradiated human chromosome on a mouse background in the Tc1 model of Down syndrome. PLoS One 8, e60482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Z., et al. , Duplication of the entire 22.9 Mb human chromosome 21 syntenic region on mouse chromosome 16 causes cardiovascular and gastrointestinal abnormalities. Hum. Mol. Genet. 16, 1359–1366 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Olson L. E., Richtsmeier J. T., Leszl J., Reeves R. H., A chromosome 21 critical region does not cause specific Down syndrome phenotypes. Science 306, 687–690 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wehrspaun C. C., Haerty W., Ponting C. P., Microglia recapitulate a hematopoietic master regulator network in the aging human frontal cortex. Neurobiol. Aging 36, 2443.e9–2443.e20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Logan T. T., Villapol S., Symes A. J., TGF-β superfamily gene expression and induction of the Runx1 transcription factor in adult neurogenic regions after brain injury. PLoS One 8, e59250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malko P., Syed Mortadza S. A., McWilliam J., Jiang L. H., TRPM2 channel in microglia as a new player in neuroinflammation associated with a spectrum of central nervous system pathologies. Front. Pharmacol. 10, 239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.So A. Y., et al. , Regulation of APC development, immune response, and autoimmunity by Bach1/HO-1 pathway in mice. Blood 120, 2428–2437 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang X., et al. , Bach1: Function, regulation, and involvement in disease. Oxid. Med. Cell. Longev. 2018, 1347969 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta I., et al. , Single-cell isoform RNA sequencing characterizes isoforms in thousands of cerebellar cells. Nat. Biotechnol, 10.1038/nbt.4259 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Joglekar A., et al. , A spatially resolved brain region- and cell type-specific isoform atlas of the postnatal mouse brain. Nat. Commun. 12, 463 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tardaguila M., et al. , Corrigendum: SQANTI: Extensive characterization of long-read transcript sequences for quality control in full-length transcriptome identification and quantification. Genome Res. 28, 1096 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wyman D., et al. , A technology-agnostic long-read analysis pipeline for transcriptome discovery and quantification. bioRxiv [Preprint] (2020). https://www.biorxiv.org/content/10.1101/672931v1 (Accessed 23 January 2020). [Google Scholar]
- 60.Rinaldi C., Wood M. J. A., Antisense oligonucleotides: The next frontier for treatment of neurological disorders. Nat. Rev. Neurol. 14, 9–21 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Deverman B. E., Ravina B. M., Bankiewicz K. S., Paul S. M., Sah D. W. Y., Gene therapy for neurological disorders: Progress and prospects. Nat. Rev. Drug Discov. 17, 641–659 (2018). [DOI] [PubMed] [Google Scholar]
- 62.de la Fuente L., et al. , tappAS: A comprehensive computational framework for the analysis of the functional impact of differential splicing. Genome Biol. 21, 119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harold D., et al. , Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 41, 1088–1093 (2009). Correction in: Nat. Genet. 45, 712 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lambert J. C., et al. ; European Alzheimer’s Disease Initiative Investigators, Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 41, 1094–1099 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Chen G. F., et al. , Amyloid beta: Structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 38, 1205–1235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rohan de Silva H. A., et al. , Cell-specific expression of beta-amyloid precursor protein isoform mRNAs and proteins in neurons and astrocytes. Brain Res. Mol. Brain Res. 47, 147–156 (1997). [DOI] [PubMed] [Google Scholar]
- 67.Mathys H., et al. , Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 21, 366–380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hickman S., Izzy S., Sen P., Morsett L., El Khoury J., Microglia in neurodegeneration. Nat. Neurosci. 21, 1359–1369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miyoshi G., et al. , Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. J. Neurosci. 30, 1582–1594 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hansen D. V., et al. , Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat. Neurosci. 16, 1576–1587 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kleschevnikov A. M., et al. , Hippocampal long-term potentiation suppressed by increased inhibition in the Ts65Dn mouse, a genetic model of Down syndrome. J. Neurosci. 24, 8153–8160 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siarey R. J., et al. , Increased synaptic depression in the Ts65Dn mouse, a model for mental retardation in Down syndrome. Neuropharmacology 38, 1917–1920 (1999). [DOI] [PubMed] [Google Scholar]
- 73.Goldberg-Stern H., et al. , Seizure frequency and characteristics in children with Down syndrome. Brain Dev. 23, 375–378 (2001). [DOI] [PubMed] [Google Scholar]
- 74.Scotti M. M., Swanson M. S., RNA mis-splicing in disease. Nat. Rev. Genet. 17, 19–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fastq files for Illumina reads and bam files for PacBio reads are available through the European Genome-Phenome Archive with accession number EGAS00001005691. Isoforms identified by long-read sequencing are available through the UCSC Genome browser:
Untargeted dataset: https://genome.ucsc.edu/s/csl022/DSND_snIsoSeq_sample.
SPP1 targeted dataset: https://genome.ucsc.edu/s/csl022/SPP1_scIsoSeq.
APP targeted dataset: https://genome.ucsc.edu/s/csl022/APP_scIsoSeq.
BIN1 targeted dataset: https://genome.ucsc.edu/s/csl022/BIN1_snIsoSeq.