Abstract
Serine/arginine-rich splicing factor 3 (SRSF3; also known as SRp20), an important member of the family of SRSFs, is abnormally expressed in tumors, resulting in aberrant splicing of hub genes, such as CD44, HER2, MDM4, Rac family small GTPase 1 and tumor protein p53. Under normal conditions, the splicing and expression of SRSF3 are strictly regulated. However, the splicing, expression and phosphorylation of SRSF3 are abnormal in tumors. SRSF3 plays important roles in the occurrence and development of tumors, including the promotion of tumorigenesis, cellular proliferation, the cell cycle and metastasis, as well as inhibition of cell senescence, apoptosis and autophagy. SRSF3-knockdown significantly inhibits the proliferation and metastatic characteristics of tumor cells. Therefore, SRSF3 may be suggested as a novel anti-tumor target. The other biological functions of SRSF3 and its regulatory mechanisms are also summarized in the current review.
Keywords: SRSF3, oncogene, splicing, RNA transport, post-transcriptional regulation
1. Introduction
Eukaryotic gene expression primarily includes the processes of transcription, splicing and translation. First, DNA is transcribed into an mRNA precursor (pre-mRNA) by RNA polymerases. Then, the pre-mRNA is spliced into mature mRNA by spliceosomes, after which the mature mRNA is translated into protein. Since multiple splicing sites are usually present in pre-mRNA, several splicing isoforms may be generated in different types of cells or biological processes by alternative splicing. These isoforms have a synergistic or antagonistic biological function, and >90% of human genes are spliced after transcription (1,2). Normal gene splicing is critical for the functions of tissues and organs (3). However, aberrant splicing of key genes can lead to various diseases, particularly the occurrence and development of tumors (4–7).
Spliceosomes usually consist of small nuclear ribonucleoproteins (snRNPs), core proteins and cofactors (8). Core proteins include two highly conserved families: i) Serine/arginine-rich splicing factors (SRSFs); and ii) heterogeneous nuclear ribonucleoproteins (hnRNPs). During splicing, the serine/arginine-rich domain (RS) of the SRSF binds to the exonic splicing enhancer (ESE) of the splicing regulatory element, facilitating the binding of snRNP to the 5′-end and the 3′-end splicing sites, which cause phosphorylation or dephosphorylation of target proteins (9,10).
At present, 12 members of the SRSF family have been identified, among which SRSF3 is the smallest; the N-terminal of SRSF3 contains an RNA recognition motif that specifically recognizes the splicing sites, and the C-terminal of SRSF3 contains a highly phosphorylated serine/arginine-rich domain (RS) region (11). SRSF3 binds to cytosine-rich motifs and promotes the inclusion of exons, especially genes encoding neutral amino acids (such as serine and threonine), which can be phosphorylated (12). SRSF3 interacts with RNA cis-acting elements in a concentration- or cell differentiation-dependent manner. Moreover, SRSF3 promotes the inclusion of exons in target genes by recruiting Yes1 associated transcriptional regulator (YTHDC1). YTHDC1 is closely associated with the nuclear localization, RNA binding ability and related splicing events of SRSF3 (13,14). SRSF3 has been discovered to be widely expressed in tumor tissues and involved in the splicing of numerous hub genes, such as CD44, HER2, MDM4, Rac family small GTPase 1 (RAC1) and tumor protein p53 (TP53) (Table I; Figs. 1 and 2) (15). Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis has demonstrated that SRSF3 is highly associated with in the PI3K-AKT signaling pathway. Consequently, SRSF3 functions as an oncogene with critical regulatory roles in tumorigenesis, proliferation, metastasis, anti-apoptosis, anti-senescence and anti-autophagy (Xiong et al, unpublished data). In addition, SRSF3 plays important roles in various biological processes, such as protein translation (16), termination of transcription (17), insulin signaling (18), mRNA transport (19) and embryogenesis (20).
Table I.
Regulators, targets and splicing events of SRSF3.
| Regulator | Target | Splicing event | Isoform | Cell line | (Refs.) |
|---|---|---|---|---|---|
| HNRNPL | SRSF3 | Skipping of exon 4 | SRSF3-FL | CAL 27 | (23) |
| PTBP1 and PTBP2 | OSCC | (46) | |||
| SLU7 | PLC, H358, and HeLa | (47) | |||
| SRSF3 | SRSF3 | Inclusion of exon 4 | SRSF3-TR | K46 | (43–45) |
| Amiodarone | HeLa | (49) | |||
| Sodium arsenite | HCT116 | (50) | |||
| circ87MARCA5 | U87MG | (51) | |||
| SRSF3 | CDCA5 | Retention of intron 1 and2 | NA | PLC | (47) |
| EIF2B5 | Retention of intron 12 | EIF2B5-TR | SQ20B | (16) | |
| PDCD4 | Retention of intron 2 | Isoform2 | SW480 | (29) | |
| CD44 | Splicing of exons 8, 9, and 10 | CD44E | HEK293 | (32,59) | |
| G6PD | Inclusion of exon 12 | G6PD-FL | HepG2 | (54) | |
| GR | Inclusion of exon 9a | GRα | MDA-MB-231 | (74,75) | |
| KLF6 | Inclusion of exon 3 | KLF6-FL | Hep3B, Huh7, and HepG2 | (22) | |
| MAP4K4 | Inclusion of exon 16 | Isoform2 and 5 | HCT-8 | (73) | |
| MDM4 | Inclusion of exon 6 | MDM4-FL | A375 | (67) | |
| NUMB | Inclusion of exon 12 | NUMB-FL | MDA-MB-231 | (76) | |
| PKM | Inclusion of exon 10 | PKM2 | DLD-1 and WiDr | (61) | |
| CASP2 | Skipping of exon 9 | Casp-2L | HeLa and MDA-MB-231 | (107) | |
| CLK1 | Skipping of exon 4 | CLK1-TR | WI-38 | (40) | |
| FoxM1 | Skipping of exon 9 and/or 6 | Isoform b and c | U2OS and HeLa | (24) | |
| HER2 | Skipping of exon 16 | Δ16HER2 | SKBR3 | (108) | |
| ELP1 | Skipping of exon 20 | ELP1-TR | HEK293 | (40) | |
| ILF3 | Skipping of exon 18 or 18b | Isoform1 and2 | WI-38 | (66) | |
| KMT2C | Skipping of exon 46a | KMT2C-S | A2780 | (68) | |
| PDK1 | Skipping of exon 3a | PDK1-TR | HEK293 | (40) | |
| RAC1 | Skipping of exon 3b | RAC1b | HT29 | (33) | |
| SMN2 | Skipping of exon 7 | SMN2-TR | HeLa | (83) | |
| MAPT | Skipping of exon 10 | MAPT-TR | SKN-MC and C33A | (84) | |
| TP53 | Skipping of exon 9i | p53β | MRC-5 | (25) | |
| TP53I3 | Skipping of exon 4 | TP53I3-TR | HEK293 | (40) |
SRSF3, serine/arginine rich splicing factor 3; NA, not applicable.
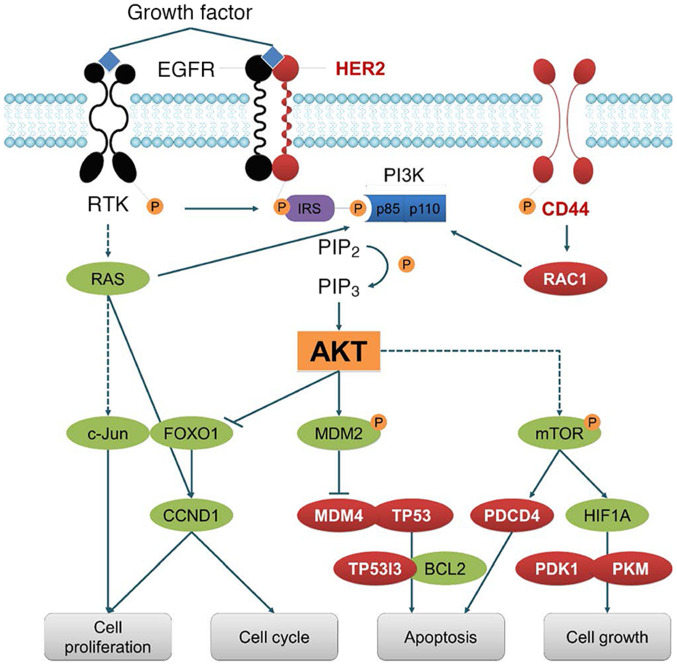
Figure 1.
Targets of SRSF3. SRSF3 is involved in the splicing of numerous hub genes, including CD44, HER2, MDM4, RAC1 and TP53. Therefore, SRSF3 plays important roles in the regulation of cell proliferation, cell cycle, apoptosis and proliferation. SRSF3, serine/arginine rich splicing factor 3; MDM, mouse double minute; RAC1, Rac family small GTPase 1; TP53, tumor protein p53; RTK, receptor tyrosine kinases; IRS, insulin receptor substrate; PIP3, phosphatidylinositol (3,4,5)-trisphosphate; PIP2, phosphatidylinositol 4,5-bisphosphate; CCND1, cyclin D1; PDCD4, programmed cell death 4; PDK1, pyruvate dehydrogenasekKinase 1; PKM, pyruvate kinase M; HIF1A, hypoxia inducible factor 1 subunit α; P, phosphate.
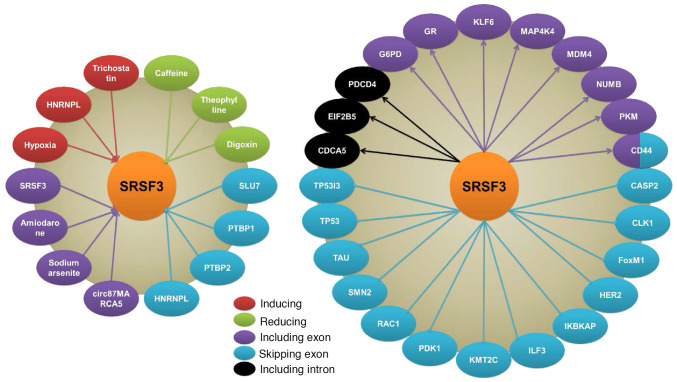
Figure 2.
Regulators, targets and splicing events of SRSF3. Hypoxia, HNRNPL and trichostatin induce SRSF3 expression, while caffeine, theophylline and digoxin inhibit SRSF3 expression. The splicing of SRSF3 is regulated by SRSF3 itself, amiodarone, sodium arsenite and cir87MARCA5 by including exon, and also regulated by HNRNPL, PTBP1, PTBP2 and SLU7 through exon skipping. SRSF3 regulates splicing of multiple genes in different ways such as including exon, skipping exon and including intron. SRSF3, serine/arginine rich splicing factor 3; HNRNPL, heterogeneous nuclear ribonucleoprotein L; PTBP, polypyrimidine tract binding protein; SLU7, SLU7 homolog.
The present review outlines the regulation of SRSF3 expression and its recently identified functions, as well as its implications in cancer. Other biological functions of SRSF3, and its regulatory mechanisms, are also summarized herein.
2. Regulatory mechanisms of SRSF3 expression
Expression of SRSF3 in cancers
SRSF3 was discovered to be aberrantly upregulated in various tumor types, including colorectal cancer (CRC) (21), gastric cancer (GC), hepatocellular carcinoma (HCC) (22), non-small cell lung cancer (NSCLC), breast cancer (BC) and oral squamous cell carcinoma (OSCC) (23). High expression of SRSF3 has also been correlated with the occurrence and development of cancer types such as CRC, bladder cancer and BC (21,24–28). Cells with high expression levels of SRSF3 demonstrate typical characteristics of tumor cells, such as accelerated cell cycle progression (21,24), enhanced anti-apoptotic ability, pro-proliferation, increased tumorigenicity (24), aerobic glycolysis (29) and anti-senescence (25). Park et al (30) detected the expression of splicing factors in GC and CRC tissues, and found that the expression rates of SRSF3 (74%) and heterogeneous nuclear ribonucleoprotein A1 (88%) were the highest in CRC tissues.
Regulation of SRSF3 expression
Mechanistic studies have revealed that hypoxia is an activator of SRSF3 in PC3 prostate cancer cells (31). Hypoxia leads to retained intron (RI)12 status of eukaryotic translation initiation factor 2B subunit ε (EIF2B5) by promoting the expression of SRSF3 and enhancing the specific binding of the RI to SRSF3 and Ser2 RNA polymerase II (16). The RI of EIF2B5 is a major regulator of translational initiation, which contains a premature termination codon. This leads to premature termination of protein translation and generates truncated EIF2B5 (EIF2B5-TR). EIF2B5-TR expression results in an increase in the survival of head and neck cancer cells during hypoxia, facilitating adaptation to the hypoxic state (16). In CRC, SRSF3 is upregulated via signaling pathways involving WNT and catenin β1 (CTNNB1)/transcription factor 4 (32–34). Allele T of single nucleotide polymorphism rs1122608 in the SMARCA4 gene was found to be associated with increased expression of SRSF3, which reduces the expression and secretion of interleukin (IL)-1β, thereby reducing the risk of atherosclerosis and stroke (35). In addition, SRSF3 expression is significantly inhibited in M. tuberculosis H37Rv-infected THP-1 macrophages (36). However, the regulatory mechanisms of the rs1122608-T allele and H37Rv in SRSF3 expression are remain unknown.
Xenobiotic regulation of SRSF3
Caffeine inhibits SRSF3 expression in HeLa cells and converts p53α to p53β to promote cell cycle arrest and apoptosis (37). Similarly, theophylline downregulates SRSF3 expression in HeLa and MCF-7 cells, and facilitates p53α-to-p53β conversion (38). Moreover, the study indicated that theophylline induces apoptosis and senescence, and reduces cell colony formation. Notably, theophylline inhibited cellular proliferation, whereas caffeine enhanced proliferation rates; neither theophylline nor caffeine had an effect on the proliferation of normal breast cells (MCF-10A). These findings suggest that theophylline and caffeine may act as antitumor compounds by targeting SRSF3 and its target genes. In addition, digoxin inhibits SRSF3 expression in HeLa cells and converts p53α to p53β isoforms, thereafter, affecting the cell cycle and DNA damage repair (39). In cells derived from patients with familial dysautonomia, digoxin exerts a therapeutic effect by inhibiting SRSF3 expression and increasing the expression of wild-type elongator complex protein 1 (ELP1) containing exon 20 (40). In addition, digoxin exerts antiviral effects by inhibiting SRSF3 expression in HIV-1, altering viral RNA splicing sites, and leading to loss of the essential viral factor Rev and the synthesis of viral structural protein (41). Trichostatin, a histone deacetylase inhibitor, acts on dendritic cells by upregulating SRSF3, hypoxia inducible factor 1 subunit α-dependent glycolytic genes and pyruvate kinase M2 (PKM2) (42). These studies offer insights into the pharmacodynamic roles of SRSF3 in cancer therapy.
Splicing of SRSF3
In the late 1990s, Jumaa and Nielsen (43–45) first demonstrated that SRSF3 regulated its own expression by promoting the inclusion of exon 4, which contains a stop codon, and consequently expresses SRSF3-TR. In OSCC cells, polypyrimidine tract binding protein (PTBP) 1 and PTBP2 bind to exonic splicing suppressors of SRSF3 exon 4 to inhibit its inclusion, which results in the upregulation of full-length SRSF3 (SRSF3-FL) (46). Heterogeneous nuclear ribonucleoprotein L has also been observed to promote the transcription and skipping of exon 4 in SRSF3 (23). A splicing regulator, SLU7, promotes exon 4 skipping in SRSF3 (47) and plays an important role in hepatocellular differentiation (48). SLU7 is also a key gene in hepatocyte homeostasis, the expression of which is inhibited in liver cancer. SLU7-knockdown in human hepatocytes results in impaired glucose and lipid metabolism, generation of key metabolic hormone tolerance, and reversal of fetal-like gene expression patterns by increasing the expression of SRSF3-TR (47,48). Amiodarone has been reported to promote the inclusion of the SRSF3 stop codon and consequent SRSF3-TR expression and apoptosis of tumor cells (49). Treatment of human colon cancer cells with sodium arsenite also increases the incidence of the SRSF3 termination codon and consequently increases SRSF3-TR expression by inhibiting nonsense-mediated mRNA decay (50). Knockout of SRSF3-TR significantly decreases the arsenite-induced JUN protein and the binding and promotive activity of the transcription factor JUN in the IL-8 promoter, thereby significantly decreasing IL-8 expression (50). Moreover, the inclusion of SRSF3 exon 4 is also induced by the overexpression of circ87MARCA5 in U87MG cells, which consequently inhibits the proliferation and migration of U87MG cells (51).
Phosphorylation of SRSF3
The localization and activity of SRSF3 are stimulated by phosphorylation of its C-terminal RS domain. The conserved SR Protein Kinase (SRPK)-specific substrate docking groove in SRPK2 affects the phosphorylation and localization of SRSF3. SRPK-mediated SRSF3 phosphorylation sites extend throughout the RS domain. In addition, hypothalamic gonadotropin-releasing hormone (GnRH) and AKT have also been identified as inducers of SRSF3 phosphorylation (52,53). Thus, inhibition of SRSF3 can reverse the GnRH-elevated Homer1a-to-Homer1b/c ratio and regulate the effect of GnRH on the expression of follicle stimulating hormone β and luteinizing hormone subunit β (52). In rat primary hepatocytes, insulin also promotes the phosphorylation of SRSF3, which in turn bonds to the ESE in exon 12 of glucose-6-phosphate dehydrogenase (G6PD), resulting in increased splicing and expression of G6PD (54). Overexpression of protein phosphatase, Mg2+/Mn2+ dependent 1G (PPM1G) promotes the dephosphorylation of SRSF3 and changes the alternative splicing patterns of genes related to the cell cycle and transcriptional regulation in HCC cells (55). SRSF3 is phosphorylated at AKT consensus sites downstream of PI3K-mediated platelet-derived growth factor receptor α signaling in mouse palatal mesenchymal cells, resulting in its nuclear translocation (56). However, SRSF3 appears to be hypo-phosphorylated and stable in cells (57), which may be a way to maintain cell homeostasis.
3. Oncogenic roles of SRSF3
Promotion of tumorigenesis
Studies have reported that SRSF3-mediated aberrant splicing is closely associated with hepatocarcinogenesis (47,58). SLU7 is a key factor in genomic stability; SLU7-knockdown results in transcription-related RNA-DNA hybrids (R-loops), DNA damage, cell cycle arrest and severe mitotic disorders, as well as loss of sister chromatid cohesion, through the production of SRSF3-TR and abnormal splicing, and expression of SRSF1 and the key sister chromatid cohesion protein cell division cycle associated 5 (47). This unique function of SLU7 has been observed in cancer cells and the livers of normal mice, demonstrating the conserved and essential role of SLU7 in preserving genomic integrity. Therefore, low expression of SLU7 in the cirrhotic liver may be involved in the development of hepatocarcinogenesis (47). RNA sequencing revealed that 50% of HCC tissues had aberrant splicing of the coiled-coil domain of coiled-coil domain containing 50 (CCDC50). Overexpression of truncated CCDC50 (CCDC50-TR) promotes HCC growth and metastasis by activating the RAS/FOXO4 signaling pathway; inhibition of MAP2K7/MAPK1 phosphorylation or overexpression of FOXO4 significantly attenuates the CCDC50-TR-mediated phenotype. SRSF3 directly binds to CCDC50-TR mRNA to maintain its stability in the cytoplasm (58). The retention of SRSF3 in the cytoplasm is mediated by the interaction between HBV-encoded X protein (HBx) and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein β (YWHAB). HBx upregulation induces the expression of SRSF3 and CCDC50-TR in the cytoplasm. Therefore, the HBx/SRSF3/YWHAB complex upregulates CCDC50-TR and promotes the progression of HCC via the RAS/FOXO4 signaling pathway (58).
The cell adhesion molecule CD44 is also a target gene of SRSF3. SRSF3 alternatively splices exons 8, 9 and 10 of CD44 to generate isoform CD44E, which promotes tumorigenesis in BC and GC (32,59). protein phosphatase 1 regulatory inhibitor subunit 1B (PPP1R1B) regulates the stability of SRSF3 protein and forms a complex with SRSF3 to regulate the expression of CD44E (60). Knockdown of PPP1R1B significantly downregulates CD44E expression in GC cells, and decreases the expression of SRSF3 and CD44E in transplanted tumors. In addition, SRSF3 is one of the major regulators of induced pluripotent stem cell reprogramming and plays an important role in dedifferentiation of tumorigenesis (61). SRSF5, a novel oncogenic splicing factor, is overexpressed in OSCC tissues and cells and is critical for the proliferation and tumorigenesis of OSCC cells (62). Overexpression of SRSF5 transforms immortal rodent fibroblasts to form tumors in nude mice, while downregulation of SRSF5 in OSCC cells impedes proliferation, cell cycle progression, and tumor growth. SRSF3 is also upregulated in OSCC tissues, especially in patients with lymphatic metastasis (62). SRSF3 induces poison cassette usage and/or intron 5 retention (63) or inhibits the usage of the proximal 3′-splice site in exon 6 of SRSF5, which disrupts SRSF5-mediated self-regulation and results in the upregulation of full-length SRSF5 in cancer cells (62). Knockdown of SRSF3 inhibits snail family transcriptional repressor 1 and cadherin 2 (CDH2) expression, while the expression of SRSF3 is induced in primary cultured oral epithelial cells by the carcinogen dimethylol butanoic acid, suggesting that SRSF3 is involved in the occurrence of oral cancer (64).
Promotion of cellular proliferation
The oncogene HER2 (erb-b2 receptor tyrosine kinase 2) is overexpressed in 20–30% of invasive BC and is associated with poor patient prognosis. SRSF3 specifically binds to HER2 exon 15 and generates the oncoprotein Δ16HER2 (with exclusion of exon 16). Downregulation of SRSF3 results in decreased expression of Δ16HER2 and increased expression of subtypes with inclusion of exon 15a or intron 15, thereby inhibiting the proliferation of BC cells (65). SRSF3 also promotes the proliferation of cancer cells by splicing interleukin enhancer binding factor 3 (ILF3), of which isoform-1 and −2 induce proliferation and transformation, whereas isoform-5 and −7 inhibit proliferation and induce apoptosis in tumor cells. SRSF3 directly binds to the RNA motif of ILF3, resulting in exclusion/inclusion of, or 3-terminal alternative splicing of, exon 18. Tumor cells with low expression of SRSF3 generate ILF3 isoform-5 and −7, thereby inhibiting the proliferation of tumor cells (66). In a variety of human cancers, SRSF3 promotes the inclusion of MDM4 exon 6 and results in increased expression of full-length MDM4, an inhibitor of p65 (67). Thus, antisense oligonucleotide-mediated MDM4 exon 6 skipping significantly inhibits MDM4 expression and exerts good antineoplastic activity in a variety of human melanoma cells and patient-derived melanoma xenografts, as well as enhancing the sensitivity of tumors to MAPK-targeting therapeutics (67). Knockdown of SRSF3 in ovarian cancer cells affects homologous recombination-mediated DNA repair activity and increases the expression of the DNA double-strand break marker H2A.X variant histone. Moreover, the splicing pattern of lysine methyltransferase 2C, an H3K4-specific histone methyltransferase, is altered, resulting in decreased expression of mono-methylated and tri-methylated H3K4 (68). SRSF3 also increases the expression of PKM2 by promoting inclusion of exon 10 in PKM2. However, SRSF3-knockout increases PKM1 expression, decreases the generation of lactic acid, and consequently inhibits cellular proliferation, suggesting that SRSF3 also promotes proliferation by regulating cancer cell metabolism (61). Various studies have indicated that microRNAs (miRNAs/miRs) are involved in the SRSF3-mediated promotion of cancer cell proliferation. For instance, knockdown of SRSF3 results in decreased RE1 silencing transcription factor and increased miR-132-3p and miR-212-3p expression, which directly inhibit Yes1 associated transcriptional regulator and subsequent cyclin D1 (CCND1), thereby suppressing the proliferation of cancer cells (69). Knockdown of SRSF3 also decreases miR-1908-5p expression by inhibiting the transactivation of NF-κB. miR-1908-5p directly inhibits NFKB inhibitor interacting Raslike 2 (NKIRAS2) expression, a negative regulator of the NF-κB pathway. Silencing NKIRAS2 inhibits NFKB inhibitor β and promotes NF-κB translocation into the nucleus, consequently activating miR-1908-5p and promoting tumor cell proliferation (70).
Promotion of the cell cycle
SRSF3 is associated with the formation of chromatin before and after mitosis, and is involved in regulation of the structure and function of chromatin, demonstrating the important roles of SRSF3 in the cell cycle (71). It has been reported that SRSF3 promotes cell cycle progression from the G2 to the M phase by regulating transcription factor forkhead box M1 and its downstream target genes, polo like kinase 1 and cell division cycle 25B (24). Therefore, inhibition of SRSF3 in tumor cells leads to cell cycle arrest (21,24). RAC1 is a small GTPase of the Rho family, which is involved in tumor cell proliferation and invasiveness by regulating actin-dependent cell motility and gene transcription. In CRC cells, SRSF3 alternatively splices exon 3b of RAC1 to generate the isoform RAC1b, so as to maintain tumor cell viability and promote tumor progression (33). Knockdown of SRSF3 in CRC cells decreases the expression of CCND1, CCND3, CCNE1, E2F1 and E2F7, and consequently prevents cell progression from G1 to S phase. Ajiro et al (59) investigated RNA splicing and gene expression regulated by SRSF3 in tumor cells at the genome-wide level, and revealed that SRSF3 regulated the expression of ≥224 genes, most of which are associated with the cell cycle or proliferation. Spliced exons are rich in two binding motifs of SRSF3, CCAGC(G)C and A(G)CAGCA. In addition, SRSF3 significantly regulates the expression of at ≥20 miRNAs, including a subset of oncogenic or tumor-suppressive miRNAs, indicating that SRSF3 maintains cellular homeostasis by affecting overall changes in gene expression.
Promotion of tumor metastasis
Studies have supported that SRSF3 facilitates the migration and invasiveness of CRC cells. Torres et al (72) detected the protein expression profile of two pairs of cell lines, KM12SM/KM12C and SW620/SW480, indicating that SRSF3 was highly expressed in metastatic KM12SM and SW620 cells. Knockout of SRSF3 significantly inhibited the adhesion, survival, proliferation, migration and liver homing abilities of KM12 cells. The observation that SRSF3 expression in CRC stem cells is significantly higher than in non-cancer stem cells further suggests the involvement of SRSF3 in metastasis (34). Lin et al (73) discovered that SRSF3 induces CRC cell metastasis through a RBM4-SRSF3-MAP4K4 axis and consequent MAPK8 signaling pathway. SRSF3 promotes the inclusion of MAP4K4 exon 16 in CRC cells, which activates the phosphorylation of MAPK8 and subsequent expression of CDH2 and vimentin. Moreover, the authors found that expression deficiency of SRSF3 was significantly associated with poor overall and disease-free-survival in patients. Previous studies have also shown that high expression of GR (NR3C1) in triple-negative breast cancer (TNBC) is associated with chemoresistance and increased mortality rates. receptor for activated C kinase 1 (RACK1) is a scaffold protein with a glucocorticoid response element site on its promoter, which participates in the migration and invasiveness of BC cells. SRSF3 alternatively splices GR to produce GRα, which regulates RACK1 expression at the transcriptional level and is involved in cell migration (74,75). In addition, SRSF3-mediated splicing of GR is positively regulated by cortisol (74,75). Clinical data indicate that TAR DNA binding protein (TARDBP) is highly expressed in TNBC and is associated with poor patient prognosis. Knockout of TARDBP inhibits the proliferation and metastasis of tumor cells, while its overexpression promotes proliferation and malignancy in mammary epithelial cells. As an important splicing regulator, TARDBP is responsible for the unique splicing profile in TNBC (76). TARDBP regulates gene splicing by interacting with SRSF3. For instance, knockdown of TARDBP or SRSF3 prevents the inclusion of NUMB endocytic adaptor protein exon 12 and thereby inhibits the metastasis and proliferation of TNBC cells (76). The intensity and prevalence of SRSF3 expression in invasive epithelial ovarian cancer cells are significantly higher than those in tumor lesions, suggesting that SRSF3 is associated with tumor cell metastasis. This was confirmed by in vitro experiments that silence SRSF3, significantly inhibiting the proliferation and metastasis abilities of U2OS and HeLa cells (77).
Inhibition of apoptosis via programmed cell death 4 (PDCD4)
It has been reported that SRSF3 promotes tumor progression by inhibiting the expression of the pro-apoptotic protein PDCD4 (60). SRSF3 inhibits alternative splicing and nuclear export of the minor isoform of PDCD4 mRNA, as well as the translation of the major isoform. Thus, the absence of PDCD4 isoforms in various cancers may be associated with the high expression of SRSF3 (78). Moreover, silencing of SRSF3 leads to a significant increase in PDCD4 mRNA in translated polysome fractions, whereas overexpression of SRSF3 induces the transformation of PDCD4 mRNA to non-translational ribosomal components (78). Another study demonstrated that SRSF3 co-localizes with PDCD4 mRNA in the cytoplasmic processing corpuscle of live cells. SRSF3 directly binds to the 5′ untranslated region (UTR) of PDCD4 and inhibits its expression (29). Moreover, the expression of anti-apoptotic proteins BCL2 and homeodomain interacting protein kinase 2 are also inhibited by SRSF3 silencing, thereby promoting the apoptosis of CRC cells (33).
Inhibition of cell senescence via p53
Senescence is the ability of cells to enter an irreversible proliferative stagnation state, and is an important mechanism for preventing cancers. Through direct binding, SRSF3 alternatively splices TP53 to produce p53β, thereby preventing p53-mediated cellular senescence (25). Down-regulation of SRSF3 in human cells promotes cellular senescence through its alternative splicing-dependent function, which is an important biological process to prevent cancer and promote individual senescence (25). Moreover, downregulation of SRSF3 induces cellular senescence through alternative polyadenylation (poly(A)) (79). Knockout of SRSF3 results in preferential use of the proximal polyadenylation site, leading to an overall shortening of the 3′-UTR of the senescence-associated mRNA. For instance, silencing of SRSF3 induces the expression of PTEN, resulting in decreased AKT phosphorylation and senescence-associated phenotypes (79). Astrocytes express both Δ133p53 and p53β isoforms, which respectively show toxicity to, and protection of, neurons. In vitro aging is caused by successive batches of primary human astrocytes. Thus, SRSF3 can be expected to be a therapeutic target for neurodegenerative diseases by reducing the expression of Δ133p53 and increasing the expression of p53β (80).
Inhibition of cell autophagy via p65 and FoxO1
Autophagy is an evolutionarily conserved cell catabolic process. Dysfunction in the autophagy pathway has been shown to be associated with numerous human disorders, including cancer (81). SRSF3 is significantly downregulated during hypoxia-induced autophagy in OSCC cells. SRSF3-knockdown induces autophagy whereas SRSF3 overexpression inhibits hypoxia-induced autophagy. Mechanistic investigation demonstrates that SRSF3 inhibits cell autophagy by suppressing the expression of p65 and FoxO1, as well as their downstream target gene beclin 1, a key regulator of autophagy (82).
4. Other functions of SRSF3
SRSF3 in non-cancerous diseases
Spinal muscular atrophy (SMA) is a major genetic factor in pediatric death and is caused by dysregulation of survival of motor neuron (SMN) proteins, of which SMN2 with exclusion of exon 7 is regulated by SRSF3 and encodes a function-deficient protein. Knockout of SRSF3 increases the inclusion of SMN2 exon 7 and the expression level of SMN protein, providing a possible therapeutic approach for SMA (83). The p53 isoforms Δ133p53 and p53β are expressed in astrocytes and regulate the toxicity and protection of neurons. SRSF3 decreases Δ133p53 and increases p53β, which significantly increases the number of senescent astrocytes and senescence-like astrocytes in the brain tissues of patients with Alzheimer's disease and amyotrophic lateral sclerosis (80). Moreover, SRSF3 promotes the development of microtubule associated protein τ (MAPT) proteinosis by inducing skipping of the MAPT exon 10 (84). SRSF3 can also cause the muscle-specific symptoms of hereditary myopathy with lactic acidosis via erroneous splicing of mutant iron-sulfur cluster assembly enzyme (85). Knockout of the Srsf3 gene in mouse hepatocytes results in destruction of the liver structure, growth retardation before and after birth, impaired hepatocyte maturation, and changes in glucose and lipid homeostasis that are manifested as decreased glycogen storage, fasting hypoglycemia, increased insulin sensitivity, and decreased cholesterol synthesis. Multiple aberrant splicing events occur in the Srsf3-knockout mouse liver, especially the aberrant splicing of key regulators of glucose and lipid metabolism, such as Hnf1α, Ern1, Hmgcs1, Dhcr7 and Scap genes (86). In addition, Srsf3 is critical to maintaining the transcriptome integrity of mouse oocytes. Conditional deletion of Srsf3 in mature germinal vesicle oocytes significantly impairs germinal vesicle breakdown, and thus oocytes are unable to enter into meiosis (87).
Interestingly, SRSF3 also regulates the translational efficiency of viral RNA by interacting with the RNA binding protein poly(rC) binding protein 2 (88). Viral mRNAs generated from intronless genes are particularly unstable owing to degradation by nuclear RNA exosomes. The viral RNA binding protein microtubule associated protein RP/EB family member 2 (MAPRE2) stabilizes viral mRNAs in the nucleus, and stimulates their export to the cytoplasm and subsequent translation into proteins. In the absence of MAPRE2, SRSF3 destabilizes viral mRNAs by interacting with RNA exosomes and their adaptor complex NEXT (89). SRSF3 also participates in the splicing of the human papillomavirus (HPV) 18 pre-mRNAs (90). If differentiated keratinocytes are infected by HPV16, E2 protein stimulates SRSF3, L1 mRNA and capsid protein expression, and thus controls late events in the viral life cycle (91). Among Kaposi's sarcoma-associated herpesviruses, SRSF3 is a cellular cofactor involved in open reading frame (ORF)57-mediated K8β splicing. In the absence of ORF57, SRSF3 binds to the K8β intron and inhibits K8β splicing, whereas knockdown of SRSF3 promotes K8β splicing. The N-terminal half of ORF57 binds to the RNA recognition motif of SRSF3, preventing the binding of SRSF3 to the K8β intron, thereby attenuating the inhibitory effect of SRSF3 on K8β splicing (92).
SRSF3 also plays a role in cardiac diseases. SRSF3 is vital for heart development and homeostasis. Cardiomyocyte-specific loss of SRSF3 expression results in decapping of critical mRNAs involved in cardiac contraction (93). Moreover, a cardiomyocyte-specific Srsf3 conditional knockout mouse model showed changes in expression levels and alternative splicing of several transcripts related to mitochondrial integrity and oxidative phosphorylation. Thus, SRSF3 also regulates mitochondrial integrity and function (94).
Involvement in miRNA maturation
The RNA-induced silencing complex (RISC), consisting of drosha ribonuclease III (DROSHA) and DGCR8 microprocessor complex subunit (DGCR8) dimer, is responsible for the splicing of primary miRNA transcripts (pri-miRNA). DROSHA and DGCR8 localize RISC by interacting with the basal UG motifs and the apical UGU motifs of the pri-miRNA, respectively. SRSF3 binds to CNNC motifs, which are located ~17 nt from the RISC binding sites, recruiting DROSHA to the basal junction, thereby enhancing RISC activity and facilitating pri-miRNA splicing (95). Fernandez et al (96) found that the genetic variation G>A in pri-mir-30c-1 results in secondary RNA structure rearrangement, which enhances the binding of SRSF3 to pri-mir-30c-1 and consequently, the level of miR-30c. The involvement of SRSF3 in miRNA processes is also supported by the finding that knockdown of SRSF3 results in increased miR-132-3p and miR-212-3p (69) and decreased miR-1908-5p (70). Moreover, Ajiro et al (59) reported that at least 20 miRNAs were dysregulated by SRSF3 in tumor cells. However, the mechanisms underlying the involvement of SRSF3 in the processing of miRNA are still unclear.
Contribution to RNA transport
Apart from splicing activities, SRSF3 is also involved in nuclear RNA export factor 1 (NXF1)-dependent RNA transport from the nucleus to the cytosol (97–99). For instance, SRSF3 directly binds to the transcript of the histone H2A gene, promoting its transport from nucleus to cytoplasm (100). YTHDC1, a recognition protein of N6-methyladenosine, is also required in the process of precursor mRNA transcripts in the oocyte nucleus through interaction with SRSF3 (101). In HeLa cells, YTHDC1 interacts with SRSF3 and promotes the binding of RNA to SRSF3 and NXF1, thereby driving export of methylated mRNA from the nucleus to the cytoplasm (102). In mice, SRSF3 binds to the core pluripotent transcription factor nanog homeobox (NANOG) mRNA and facilitates its transport from the nucleus to the cytoplasm. In the absence of SRSF3, NANOG mRNA is sequestered in the nucleus and its protein expression level is markedly reduced. In addition, SRSF3 also regulates alternative splicing of the translocator Nxf1 and RNA regulators, as well as the steady-state level of mRNA that encodes chromatin modifications (19). SRSF3 promotes the binding of NXF1 to mRNA and confers the sequence specificity of NXF1 to the last exon of that RNA. Thus, the alternative splicing and poly(A) is linked with NXF1-mediated mRNA transport through SRSF3, which ensures the abundance of transcripts with different 3′-UTR lengths in the cytoplasm (103).
Post-transcriptional regulation of gene expression
SRSF3 is an important component of RNA granules, such as stress granules and cytoplasmic processing corpuscles, and plays an important role in post-transcriptional regulation in the cytoplasm (104). Brugiolo et al (17) determined the binding sites of SRSF3 on target mRNAs and RNAs in chromatin, the nucleoplasm and cytoplasmic subcellular fractions. The authors found that SRSF3 bonded to RNA in both the nucleus and cytoplasm. SRSF3 also participates in the poly(A) of mRNA, indicating its role in recognizing terminal exons in mRNA precursor splicing (105). SRSF3 modulates the recognition of poly(A) splicing sites in calcitonin-encoding gene calcitonin related polypeptide α by disrupting the binding of cleavage stimulation factor subunit 2 (105). In addition, Boutej et al (106) found that SRSF3 inhibited the translation of overexpressed mRNA by binding to its 3′-UTR. Serum amyloid A3, and other genes that are regulated by lipopolysaccharide, contain multiple binding sites for SRSF3 in their 3′-UTRs. By inhibiting the translation of lipopolysaccharide-stimulated genes, SRSF3 acts as a major regulator of the innate immune responses in resident microglia.
5. Conclusions
SRSF3 is involved in the splicing of a variety of key genes, and has versatile biological functions. SRSF3 promotes the inclusion of exons of target mRNAs by binding to their ESE. The activity of SRSF3 depends on the phosphorylation of its C-terminal RS domain, which is regulated by SRPK, GnRH, PPM1G and insulin. Abnormal expression of SRSF3 leads to aberrant gene splicing and the subsequent occurrence and development of various diseases, such as cancers, SMA, Alzheimer's disease, amyotrophic lateral sclerosis, MAPT proteinosis and hereditary myopathy with lactic acidosis. Studies have identified abnormally highly expressed SRSF3 in various tumor tissues, including CRC, GC, HCC, NSCLC, OSCC and BC. SRSF3 expression is regulated by the WNT pathway, SLU7, and hypoxia. The expression of SRSF3 is inhibited by xenobiotics, such as caffeine, theophylline, digoxin and amiodarone. SRSF3 participates in the occurrence of tumors and promotes the proliferation and metastasis capacity of tumor cells by inhibiting apoptosis, senescence and autophagy, as well as promoting cellular proliferation and cell cycle. Progression. Furthermore, SRSF3 promotes the invasiveness and metastasis of tumor cells. In addition, SRSF3 is involved in biological processes such as RNA transport, miRNA maturation, protein translation, heart development and embryogenesis. Notably, several controversial functions of SRSF3 complicate its pathological roles. For instance, contrary to the anti-apoptosis role of SRSF3, it binds to exon 8 of caspase 2 (CASP2) pre-mRNA, promoting the skipping of exon 9 and upregulating pro-apoptotic protein Casp-2L. Knockdown of SRSF3 induces the inclusion of exon 9 and increases anti-apoptotic protein Casp-2S (107). SRSF3 has also been reported to inhibit fibrosis by regulating gene splicing, mitotic splicing, and epithelial-mesenchymal transition (EMT) to prevent the development of HCC (18). Inhibition of SRSF3 increases the expression of insulin like growth factor and INSR and aberrantly activates the mitotic signal and promotes aberrant splicing and the expression of EMT genes. Moreover, the WNT/CTNNB1 signal pathway is activated and MYC expression is enhanced upon SRSF3 inhibition (18). Past studies suggest that the regulatory roles of SRSF3 are cell-dependent and/or disease-dependent. The function of SRSF3 in different tissue types and diseases requires further elucidation. Antisense oligonucleotide-mediated downregulation of SRSF3 makes OSCC and breast cancer cells more sensitive to the chemotherapy drug paclitaxel. Modulation of RNA splicing profiles in disease by antisense oligonucleotides is another exciting therapeutic approach that requires further investigation.
In conclusion, the present review provides an overview of the expression, regulation and function of SRSF3, as well as its mechanisms, especially the oncogenic roles. Since numerous studies have confirmed the inhibitory role of SRSF3-knockdown in tumor growth and metastasis, there is great interest in developing SRSF3 inhibitors for cancer therapy. Studying the molecular mechanisms of SRSF3-dependent signal transduction is expected to reveal additional candidates and therapeutic targets for cancer therapy.
Acknowledgements
Not applicable.
Funding Statement
Funding: The present study was supported by the National Natural Science Foundation of China (grant no. 81773044), the Science and Technology Special Project of Clinical Medicine in Jiangsu Province (grant no. BL2014046) and the Social Development Project of Jiangsu Province (grant no. BE2019657).
Availability of data and materials
Not applicable.
Authors' contributions
JX and YC collected the literature, wrote and revised the manuscript. WW and JS reviewed and revised the manuscript. All authors have read and approved the final manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baralle F, Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol. 2017;18:437–451. doi: 10.1038/nrm.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 4.Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Manley JL. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 2013;3:1228–1237. doi: 10.1158/2159-8290.CD-13-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Weiss WA. Alternative splicing in cancer: Implications for biology and therapy. Oncogene. 2015;34:1–14. doi: 10.1038/onc.2013.570. [DOI] [PubMed] [Google Scholar]
- 7.Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33:5311–5318. doi: 10.1038/onc.2013.533. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 9.Alzhanova-Ericsson AT, Sun X, Visa N, Kiseleva E, Wurtz T, Daneholt B. A protein of the SR family of splicing factors binds extensively to exonic Balbiani ring pre-mRNA and accompanies the RNA from the gene to the nuclear pore. Genes Dev. 1996;10:2881–2893. doi: 10.1101/gad.10.22.2881. [DOI] [PubMed] [Google Scholar]
- 10.Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol. 2006;13:973–980. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- 12.Fontrodona N, Aubé F, Claude JB, Polvèche H, Lemaire S, Tranchevent LC, Modolo L, Mortreux F, Bourgeois CF, Auboeuf D. Interplay between coding and exonic splicing regulatory sequences. Genome Res. 2019;29:711–722. doi: 10.1101/gr.241315.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Ye F, Chen ER, Nilsen TW. Kaposi's sarcoma-associated herpesvirus utilizes and manipulates RNA N 6-adenosine methylation to promote lytic replication. J Virol. 2017;91:e00466–e00417. doi: 10.1128/JVI.00466-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbo C, Orru S, Salvatore F. SRp20: An overview of its role in human diseases. Biochem Biophys Res Commun. 2013;436:1–5. doi: 10.1016/j.bbrc.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 16.Brady LK, Wang H, Radens CM, Bi Y, Radovich M, Maity A, CIvan C, Ivan M, Barash Y, Koumenis C. Transcriptome analysis of hypoxic cancer cells uncovers intron retention in EIF2B5 as a mechanism to inhibit translation. PLoS Biol. 2017;15:e2002623. doi: 10.1371/journal.pbio.2002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brugiolo M, Botti V, Liu N, Müller-McNicoll M, Neugebauer KM. Fractionation iCLIP detects persistent SR protein binding to conserved, retained introns in chromatin, nucleoplasm and cytoplasm. Nucleic Acids Res. 2017;45:10452–10465. doi: 10.1093/nar/gkx671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sen S, Langiewicz M, Jumaa H, Webster NJ. Deletion of serine/arginine-rich splicing factor 3 in hepatocytes predisposes to hepatocellular carcinoma in mice. Hepatology. 2015;61:171–183. doi: 10.1002/hep.27380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ratnadiwakara M, Archer SK, Dent CI, Mozos IR, Beilharz TH, Knaupp AS, Nefzger CM, Polo JM, Anko ML. SRSF3 promotes pluripotency through Nanog mRNA export and coordination of the pluripotency gene expression program. Elife. 2018;7:e37419. doi: 10.7554/eLife.37419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jumaa H, Wei G, Nielsen PJ. Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr Biol. 1999;9:899–902. doi: 10.1016/S0960-9822(99)80394-7. [DOI] [PubMed] [Google Scholar]
- 21.Kurokawa K, Akaike Y, Masuda K, Kuwano Y, Nishida K, Yamagishi N, Kajita K, Tanahashi T, Rokutan K. Downregulation of serine/arginine-rich splicing factor 3 induces G1 cell cycle arrest and apoptosis in colon cancer cells. Oncogene. 2014;33:1407–1417. doi: 10.1038/onc.2013.86. [DOI] [PubMed] [Google Scholar]
- 22.Yea S, Narla G, Zhao X, Garg R, Tal-Kremer S, Hod E, Villanueva A, Loke J, Tarocchi M, Akita K, et al. Ras promotes growth by alternative splicing-mediated inactivation of the KLF6 tumor suppressor in hepatocellular carcinoma. Gastroenterology. 2008;134:1521–1531. doi: 10.1053/j.gastro.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia R, Zhang S, Liu M, Zhang Y, Liu Y, Fan M, Guo J. HnRNP L is important for the expression of oncogene SRSF3 and oncogenic potential of oral squamous cell carcinoma cells. Sci Rep. 2016;6:35976. doi: 10.1038/srep35976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia R, Li C, McCoy JP, Deng CX, Zheng ZM. SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int J Biol Sci. 2010;6:806–826. doi: 10.7150/ijbs.6.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Y, Horikawa I, Ajiro M, Robles AI, Fujita K, Mondal AM, Stauffer JK, Zheng ZM, Harris CC. Downregulation of splicing factor SRSF3 induces p53beta, an alternatively spliced isoform of p53 that promotes cellular senescence. Oncogene. 2013;32:2792–2798. doi: 10.1038/onc.2012.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karthikeyan C, Amawi H, Viana AG, Sanglard L, Hussein N, Saddler M, Ashby CR, Jr, Moorthy NS, Trivedi P, Tiwari AK. lH-Pyrazolo[3,4-b]quinolin-3-amine derivatives inhibit growth of colon cancer cells via apoptosis and sub G1 cell cycle arrest. Bioorg Med Chem Lett. 2018;28:2244–2249. doi: 10.1016/j.bmcl.2018.05.045. [DOI] [PubMed] [Google Scholar]
- 27.Kim WJ, Lee SJ, Choi YD, Moon SK. Decursin inhibits growth of human bladder and colon cancer cells via apoptosis, G1-phase cell cycle arrest and extracellular signal-regulated kinase activation. Int J Mol Med. 2010;25:635–641. doi: 10.3892/ijmm_00000386. [DOI] [PubMed] [Google Scholar]
- 28.Morley KL, Ferguson PJ, Koropatnick J. Tangeretin and nobiletin induce G1 cell cycle arrest but not apoptosis in human breast and colon cancer cells. Cancer Lett. 2007;251:168–178. doi: 10.1016/j.canlet.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Park RY, Chen JK, Kim J, Jeong S, Ohn T. Splicing factor SRSF3 represses the translation of programmed cell death 4 mRNA by associating with the 5′-UTR region. Cell Death Differ. 2014;21:481–490. doi: 10.1038/cdd.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park WC, Kim HR, Kang DB, Ryu JS, Choi KH, Lee GO, Yun KJ, Kim KY, Park R, Yoon KH, et al. Comparative expression patterns and diagnostic efficacies of SR splicing factors and HNRNPA1 in gastric and colorectal cancer. BMC Cancer. 2016;16:358. doi: 10.1186/s12885-016-2387-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowler E, Porazinski S, Uzor S, Thibault P, Durand M, Lapointe E, Rouschop KM, Hancock J, Wilson I, Ladomery M. Hypoxia leads to significant changes in alternative splicing and elevated expression of CLK splice factor kinases in PC3 prostate cancer cells. BMC Cancer. 2018;18:355. doi: 10.1186/s12885-018-4227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goncalves V, Matos P, Jordan P. The beta-catenin/TCF4 pathway modifies alternative splicing through modulation of SRp20 expression. RNA. 2008;14:2538–2549. doi: 10.1261/rna.1253408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goncalves V, Matos P, Jordan P. Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and wnt pathways. Hum Mol Genet. 2009;18:3696–3707. doi: 10.1093/hmg/ddp317. [DOI] [PubMed] [Google Scholar]
- 34.Corbo C, Orrù S, Gemei M, Noto RD, Mirabelli P, Imperlini E, Ruoppolo M, Vecchio LD, Salvatore F. Protein cross-talk in CD133+ colon cancer cells indicates activation of the Wnt pathway and upregulation of SRp20 that is potentially involved in tumorigenicity. Proteomics. 2012;12:2045–2059. doi: 10.1002/pmic.201100370. [DOI] [PubMed] [Google Scholar]
- 35.Xiong X, Xu C, Zhang Y, Li X, Wang B, Wang F, Yang Q, Wang D, Wang X, Li S, et al. BRG1 variant rs1122608 on chromosome 19p13.2 confers protection against stroke and regulates expression of pre-mRNA-splicing factor SFRS3. Hum Genet. 2014;133:499–508. doi: 10.1007/s00439-013-1389-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang W, Niu C, Fu RY, Peng ZY. Mycobacterium tuberculosis H37Rv infection regulates alternative splicing in macrophages. Bioengineered. 2018;9:203–208. doi: 10.1080/21655979.2017.1387692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu GY, Huang SM, Liu ST, Liu PY, Chou WY, Lin WS. Caffeine induces tumor cytotoxicity via the regulation of alternative splicing in subsets of cancer-associated genes. Int J Biochem Cell Biol. 2014;47:83–92. doi: 10.1016/j.biocel.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Chang YL, Hsu YJ, Chen Y, Wang YW, Huang SM. Theophylline exhibits anti-cancer activity via suppressing SRSF3 in cervical and breast cancer cell lines. Oncotarget. 2017;8:101461–101474. doi: 10.18632/oncotarget.21464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu GY, Liu ST, Huang SM, Chang YL, Lin WS. Multiple effects of digoxin on subsets of cancer-associated genes through the alternative splicing pathway. Biochimie. 2014;106:131–139. doi: 10.1016/j.biochi.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 40.Liu B, Anderson SL, Qiu J, Rubin BY. Cardiac glycosides correct aberrant splicing of IKBKAP-encoded mRNA in familial dysautonomia derived cells by suppressing expression of SRSF3. FEBS J. 2013;280:3632–3646. doi: 10.1111/febs.12355. [DOI] [PubMed] [Google Scholar]
- 41.Wong RW, Balachandran A, Ostrowski MA, Cochrane A. Digoxin suppresses HIV-1 replication by altering viral RNA processing. PLoS Pathog. 2013;9:e1003241. doi: 10.1371/journal.ppat.1003241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang H, Zhang S, Song T, Guan X, Zhang R, Chen X. Trichostatin a protects dendritic cells against oxygen-glucose deprivation via the SRSF3/PKM2/glycolytic pathway. Front Pharmacol. 2018;9:612. doi: 10.3389/fphar.2018.00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jumaa H, Guenet JL, Nielsen PJ. Regulated expression and RNA processing of transcripts from the Srp20 splicing factor gene during the cell cycle. Mol Cell Biol. 1997;17:3116–3124. doi: 10.1128/MCB.17.6.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jumaa H, Nielsen PJ. Regulation of SRp20 exon 4 splicing. Biochim Biophys Acta. 2000;1494:137–143. doi: 10.1016/S0167-4781(00)00233-5. [DOI] [PubMed] [Google Scholar]
- 45.Jumaa H, Nielsen PJ. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo J, Jia J, Jia R. PTBP1 and PTBP2 impaired autoregulation of SRSF3 in cancer cells. Sci Rep. 2015;5:14548. doi: 10.1038/srep14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jimenez M, Urtasun R, Elizalde M, Azkona M, Latasa MU, Uriarte I, Arechederra M, Alignani D, Bárcena-Varela M, Álvarez-Sola G, et al. Splicing events in the control of genome integrity: Role of SLU7 and truncated SRSF3 proteins. Nucleic Acids Res. 2019;47:3450–3466. doi: 10.1093/nar/gkz014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elizalde M, Urtasun R, Azkona M, Latasa MU, Goñi S, García-Irigoyen O, Uriarte I, Segura V, Collantes M, Scala MD, et al. Splicing regulator SLU7 is essential for maintaining liver homeostasis. J Clin Invest. 2014;124:2909–2920. doi: 10.1172/JCI74382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang YL, Liu ST, Wang YW, Lin WS, Huang SM. Amiodarone promotes cancer cell death through elevated truncated SRSF3 and downregulation of miR-224. Oncotarget. 2018;9:13390–13406. doi: 10.18632/oncotarget.24385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kano S, Nishida K, Kurebe H, Nishiyama C, Kita K, Akaike Y, Kajita K, Kurokawa K, Masuda K, Kuwano Y, et al. Oxidative stress-inducible truncated serine/arginine-rich splicing factor 3 regulates interleukin-8 production in human colon cancer cells. Am J Physiol Cell Physiol. 2014;306:C250–C262. doi: 10.1152/ajpcell.00091.2013. [DOI] [PubMed] [Google Scholar]
- 51.Barbagallo D, Caponnetto A, Cirnigliaro M, Brex D, Barbagallo C, D'Angeli F, Morrone A, Caltabiano R, Barbagallo GM, Ragusa M, et al. CircSMARCA5 inhibits migration of glioblastoma multiforme cells by regulating a molecular axis involving splicing factors SRSF1/SRSF3/PTB. Int J Mol Sci. 2018;19:480. doi: 10.3390/ijms19020480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Q, Chikina MD, Pincas H, Sealfon SC. Homer1 alternative splicing is regulated by gonadotropin-releasing hormone and modulates gonadotropin gene expression. Mol Cell Biol. 2014;34:1747–1756. doi: 10.1128/MCB.01401-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blaustein M, Pelisch F, Tanos T, Muñoz MJ, Wengier D, Quadrana L, Sanford JR, Muschietti JP, Kornblihtt AR, Cáceres JF, et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat Struct Mol Biol. 2005;12:1037–1044. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- 54.Walsh CM, Suchanek AL, Cyphert TJ, Kohan AB, Szeszel-Fedorowicz W, Salati LM. Serine arginine splicing factor 3 is involved in enhanced splicing of glucose-6-phosphate dehydrogenase RNA in response to nutrients and hormones in liver. J Biol Chem. 2013;288:2816–2828. doi: 10.1074/jbc.M112.410803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen D, Zhao Z, Chen L, Li Q, Zou J, Liu S. PPM1G promotes the progression of hepatocellular carcinoma via phosphorylation regulation of alternative splicing protein SRSF3. Cell Death Dis. 2021;12:722. doi: 10.1038/s41419-021-04013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dennison B, Larson ED, Fu R, Mo J, Fantauzzo KA. Srsf3 mediates alternative RNA splicing downstream of PDGFRα signaling in the facial mesenchyme. Development. 2021;148:dev199448. doi: 10.1242/dev.199448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Long Y, Sou WH, Yung KW, Liu H, Wan SW, Li Q, Zeng C, Law CO, Chan GH, Lau TC, Ngo JC. Distinct mechanisms govern the phosphorylation of different SR protein splicing factors. J Biol Chem. 2019;294:1312–1327. doi: 10.1074/jbc.RA118.003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen H, Gao F, He M, Ding XF, Wong AM, Sze SC, Yu AC, Sun T, Chan AWH, Wang X, Wong N. Long-read RNA sequencing identifies alternative splice variants in hepatocellular carcinoma and tumor-specific isoforms. Hepatology. 2019;70:1011–1025. doi: 10.1002/hep.30500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ajiro M, Jia R, Yang Y, Zhu J, Zheng Z. A genome landscape of SRSF3-regulated splicing events and gene expression in human osteosarcoma U2OS cells. Nucleic Acids Res. 2016;44:1854–1870. doi: 10.1093/nar/gkv1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu S, Chen Z, Katsha A, Hong J, Belkhiri A, El-Rifai W. Regulation of CD44E by DARPP-32-dependent activation of SRp20 splicing factor in gastric tumorigenesis. Oncogene. 2016;35:1847–1856. doi: 10.1038/onc.2015.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuranaga Y, Sugito N, Shinohara H, Tsujino T, Taniguchi K, Komura K, Ito Y, Soga T, Akao Y. SRSF3, a splicer of the PKM gene, regulates cell growth and maintenance of cancer-specific energy metabolism in colon cancer cells. Int J Mol Sci. 2018;19:3012. doi: 10.3390/ijms19103012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang S, Jia R, Bian Z. SRSF5 functions as a novel oncogenic splicing factor and is upregulated by oncogene SRSF3 in oral squamous cell carcinoma. Biochim Biophys Acta Mol Cell Res. 2018;1865:1161–1172. doi: 10.1016/j.bbamcr.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 63.Änkö ML, Müller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13:R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peiqi L, Zhaozhong G, Yaotian Y, Jun J, Jihua G, Rong J. Expression of SRSF3 is correlated with carcinogenesis and progression of oral squamous cell carcinoma. Int J Med Sci. 2016;13:533–539. doi: 10.7150/ijms.14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gautrey H, Jackson C, Dittrich AL, Browell D, Lennard T, Tyson-Capper A. SRSF3 and hnRNP H1 regulate a splicing hotspot of HER2 in breast cancer cells. RNA Biol. 2015;12:1139–1151. doi: 10.1080/15476286.2015.1076610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jia R, Ajiro M, Yu L, McCoy P, Jr, Zheng ZM. Oncogenic splicing factor SRSF3 regulates ILF3 alternative splicing to promote cancer cell proliferation and transformation. RNA. 2019;25:630–644. doi: 10.1261/rna.068619.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dewaele M, Tabaglio T, Willekens K, Bezzi M, Teo SX, Low DH, Koh CM, Rambow F, Fiers M, Rogiers A, et al. Antisense oligonucleotide-mediated MDM4 exon 6 skipping impairs tumor growth. J Clin Invest. 2016;126:68–84. doi: 10.1172/JCI82534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He X, Zhang P. Serine/arginine-rich splicing factor 3 (SRSF3) regulates homologous recombination-mediated DNA repair. Mol Cancer. 2015;14:158. doi: 10.1186/s12943-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim HR, Hwang SJ, Shin CH, Choi KH, Ohn T, Kim HH. SRSF3-regulated miR-132/212 controls cell migration and invasion by targeting YAP1. Exp Cell Res. 2017;358:161–170. doi: 10.1016/j.yexcr.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 70.Kim HR, Shin CH, Lee H, Choi KH, Nam DH, Ohn T, Kim HH. MicroRNA-1908-5p contributes to the oncogenic function of the splicing factor SRSF3. Oncotarget. 2017;8:8342–8355. doi: 10.18632/oncotarget.14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, Macfarlan T, Manley JL, Chakravarti D. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33:450–461. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Torres S, García-Palmero I, Marín-Vicente C, Bartolomé RA, Calviño E, Fernández-Aceñero MJ, Casal JI. Proteomic characterization of transcription and splicing factors associated with a metastatic phenotype in colorectal cancer. J Proteome Res. 2018;17:252–264. doi: 10.1021/acs.jproteome.7b00548. [DOI] [PubMed] [Google Scholar]
- 73.Lin JC, Lee YC, Tan TH, Liang YC, Chuang HC, Fann YC, Johnson KR, Lin YJ. RBM4-SRSF3-MAP4K4 splicing cascade modulates the metastatic signature of colorectal cancer cell. Biochim Biophys Acta Mol Cell Res. 2018;1865:259–272. doi: 10.1016/j.bbamcr.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 74.Buoso E, Ronfani M, Galasso M, Ventura D, Corsini E, Racchi M. Cortisol-induced SRSF3 expression promotes GR splicing, RACK1 expression and breast cancer cells migration. Pharmacol Res. 2019;143:17–26. doi: 10.1016/j.phrs.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 75.Buoso E, Galasso M, Ronfani M, Serafini MM, Lanni C, Corsini E, Racchi M. Role of spliceosome proteins in the regulation of glucocorticoid receptor isoforms by cortisol and dehydroepiandrosterone. Pharmacol Res. 2017;120:180–187. doi: 10.1016/j.phrs.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 76.Ke H, Zhao L, Zhang H, Feng X, Xu H, Hao J, Wang S, Yang Q, Zou L, Su X, et al. Loss of TDP43 inhibits progression of triple-negative breast cancer in coordination with SRSF3. Proc Natl Acad Sci USA. 2018;115:E3426–E3435. doi: 10.1073/pnas.1714573115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He X, Arslan AD, Pool MD, Ho TT, Darcy KM, Coon JS, Beck WT. Knockdown of splicing factor SRp20 causes apoptosis in ovarian cancer cells and its expression is associated with malignancy of epithelial ovarian cancer. Oncogene. 2011;30:356–365. doi: 10.1038/onc.2010.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Park SK, Jeong S. SRSF3 represses the expression of PDCD4 protein by coordinated regulation of alternative splicing, export and translation. Biochem Biophys Res Commun. 2016;470:431–438. doi: 10.1016/j.bbrc.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 79.Shen T, Li H, Song Y, Li L, Lin J, Wei G, Ni T. Alternative polyadenylation dependent function of splicing factor SRSF3 contributes to cellular senescence. Aging (Albany NY) 2019;11:1356–1388. doi: 10.18632/aging.101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Turnquist C, Horikawa I, Foran E, Major EO, Vojtesek B, Lane DP, Lu X, Harris BT, Harris CC. p53 isoforms regulate astrocyte-mediated neuroprotection and neurodegeneration. Cell Death Differ. 2016;23:1515–1528. doi: 10.1038/cdd.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Levy J, Towers C, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou L, Guo J, Jia R. Oncogene SRSF3 suppresses autophagy via inhibiting BECN1 expression. Biochem Biophys Res Commun. 2019;509:966–972. doi: 10.1016/j.bbrc.2019.01.048. [DOI] [PubMed] [Google Scholar]
- 83.Wee CD, Havens MA, Jodelka FM, Hastings ML. Targeting SR proteins improves SMN expression in spinal muscular atrophy cells. PLoS One. 2014;9:e115205. doi: 10.1371/journal.pone.0115205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu Q, Guo J, Zhou J. A minimal length between tau exon 10 and 11 is required for correct splicing of exon 10. J Neurochem. 2004;90:164–172. doi: 10.1111/j.1471-4159.2004.02477.x. [DOI] [PubMed] [Google Scholar]
- 85.Rawcliffe DF, Österman L, Lindsten H, Holmberg M. The high level of aberrant splicing of ISCU in slow-twitch muscle may involve the splicing factor SRSF3. PLoS One. 2016;11:e0165453. doi: 10.1371/journal.pone.0165453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sen S, Jumaa H, Webster NJ. Splicing factor SRSF3 is crucial for hepatocyte differentiation and metabolic function. Nat Commun. 2013;4:1336. doi: 10.1038/ncomms2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Do DV, Strauss B, Cukuroglu E, Macaulay I, Wee KB, Hu TX, Igor RD, Lee C, Harrison A, Butler R, et al. SRSF3 maintains transcriptome integrity in oocytes by regulation of alternative splicing and transposable elements. Cell Discov. 2018;4:33. doi: 10.1038/s41421-018-0032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bedard KM, Daijogo S, Semler BL. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007;26:459–467. doi: 10.1038/sj.emboj.7601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mure F, Corbin A, Benbahouche NE, Bertrand E, Manet E, Gruffat H. The splicing factor SRSF3 is functionally connected to the nuclear RNA exosome for intronless mRNA decay. Sci Rep. 2018;8:12901. doi: 10.1038/s41598-018-31078-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ajiro M, Tang S, Doorbar J, Zheng ZM. Serine/arginine-rich splicing factor 3 and heterogeneous nuclear ribonucleoprotein A1 regulate alternative rna splicing and gene expression of human papillomavirus 18 through two functionally distinguishable cis elements. J Virol. 2016;90:9138–9152. doi: 10.1128/JVI.00965-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Klymenko T, Hernandez-Lopez H, MacDonald AI, Bodily JM, Graham SV. Human papillomavirus E2 regulates SRSF3 (SRp20) to promote capsid protein expression in infected differentiated keratinocytes. J Virol. 2016;90:5047–5058. doi: 10.1128/JVI.03073-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Majerciak V, Lu M, Li X, Zheng ZM. Attenuation of the suppressive activity of cellular splicing factor SRSF3 by Kaposi sarcoma-associated herpesvirus ORF57 protein is required for RNA splicing. RNA. 2014;20:1747–1758. doi: 10.1261/rna.045500.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ortiz-Sánchez P, Villalba-Orero M, López-Olañeta MM, Larrasa-Alonso J, Sánchez-Cabo F, Martí-Gómez C, Camafeita E, Gómez-Salinero JM, Ramos-Hernández L, Nielsen PJ, et al. Loss of SRSF3 in cardiomyocytes leads to decapping of contraction-related mRNAs and severe systolic dysfunction. Circ Res. 2019;125:170–183. doi: 10.1161/CIRCRESAHA.118.314515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dumont A, Dumont L, Zhou D, Giguère H, Pileggi C, Harper ME, Blondin DP, Scott MS, Auger-Messier M. Cardiomyocyte-specific Srsf3 deletion reveals a mitochondrial regulatory role. FASEB J. 2021;35:e21544. doi: 10.1096/fj.202002293RR. [DOI] [PubMed] [Google Scholar]
- 95.Kim K, Nguyen TD, Li S, Nguyen TA. SRSF3 recruits DROSHA to the basal junction of primary microRNAs. RNA. 2018;24:892–898. doi: 10.1261/rna.065862.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fernandez N, Cordiner RA, Young RS, Hug N, Macias S, Cáceres JF. Genetic variation and RNA structure regulate microRNA biogenesis. Nat Commun. 2017;8:15114. doi: 10.1038/ncomms15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang Y, Gattoni R, Stévenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–843. doi: 10.1016/S1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 98.Hautbergue GM, Hung ML, Golovanov AP, Lian LY, Wilson SA. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc Natl Acad Sci USA. 2008;105:5154–5159. doi: 10.1073/pnas.0709167105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Escudero-Paunetto L, Li L, Hernandez FP, Sandri-Goldin RM. SR proteins SRp20 and 9G8 contribute to efficient export of herpes simplex virus 1 mRNAs. Virology. 2010;401:155–164. doi: 10.1016/j.virol.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell. 2001;7:899–905. doi: 10.1016/S1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 101.Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, Schultz RM, Wang PJ. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018;14:e1007412. doi: 10.1371/journal.pgen.1007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife. 2017;6:e31311. doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Muller-McNicoll M, Botti V, de Jesus Domingues AM, Brandl H, Schwich OD, Steiner MC, Curk T, Poser I, Zarnack K, Neugebauer KM. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016;30:553–566. doi: 10.1101/gad.276477.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yoon SP, Kim HH, Kim J, Park RY, Ohn T. Regulation of cellular RNA nano-particle assembly by splicing factor SRp20. J Nanosci Nanotechnol. 2013;13:184–187. doi: 10.1166/jnn.2013.6951. [DOI] [PubMed] [Google Scholar]
- 105.Lou H, Neugebauer KM, Gagel RF, Berget SM. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol Cell Biol. 1998;18:4977–4985. doi: 10.1128/MCB.18.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Boutej H, Rahimian R, Thammisetty SS, Béland LC, Lalancette-Hébert M, Kriz J. Diverging mRNA and protein networks in activated microglia reveal SRSF3 suppresses translation of highly upregulated innate immune transcripts. Cell Rep. 2017;21:3220–3233. doi: 10.1016/j.celrep.2017.11.058. [DOI] [PubMed] [Google Scholar]
- 107.Jang HN, Lee M, Loh TJ, Choi SW, Oh HK, Moon H, Cho S, Hong SE, Kim DH, Sheng Z, et al. Exon 9 skipping of apoptotic caspase-2 pre-mRNA is promoted by SRSF3 through interaction with exon 8. Biochim Biophys Acta. 2014;1839:25–32. doi: 10.1016/j.bbagrm.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stickeler E, Kittrell F, Medina D, Berget SM. Stage-specific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene. 1999;18:3574–3578. doi: 10.1038/sj.onc.1202671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.