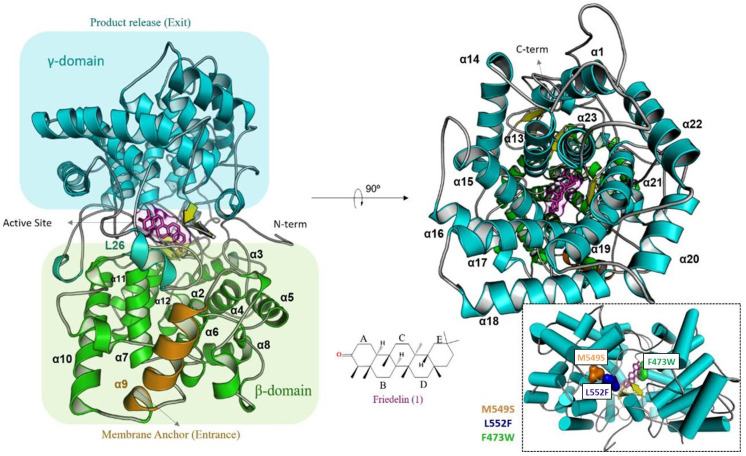
Figure 3.
In silico model of the enzyme friedelin synthase from M. ilicifolia (MiFRS). Secondary structure representation shows the α/α domains known as βγ-protein. The α9 helix approaches the lipid bilayer which sources the substrate. On the other side, the γ-domain processes and release the friedelin product. Horizontally to the N-terminal is organized the active site, created in interface between γ and β domains. Specifically, flexible loops and three β-hairpins give motion and polarity for the substrate production. Mutations made are located in loop regions as follows: L23 (Phe473Trp), L26 (Leu552Phe) and (Met549Ser). Native amino acids overview is highlighted by surfaces in colors on their positions inside the folding.