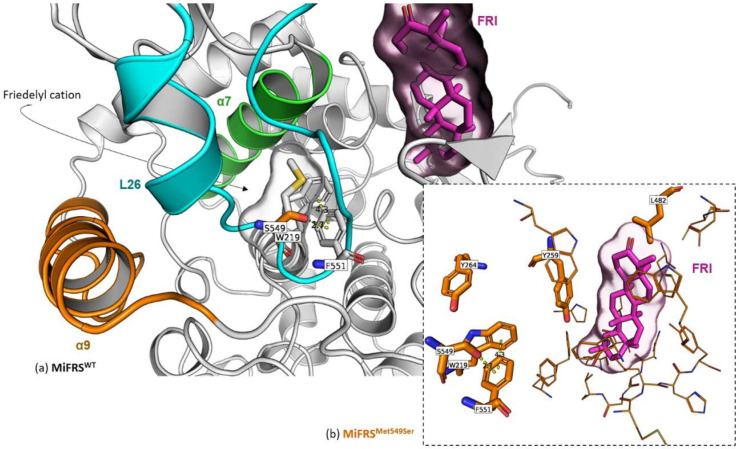
Figure 6.
Secondary alignment (or side-by-side representation) of the wild protein (MiFRS) (grey stick carbons) with MiFRSMet549Ser mutant (orange stick carbons). The surface highlights the volume adopted by the friedelin molecule in the catalytic cavity. Colored cartoons represent important protein regions: i. membrane contact α9 (orange), ii. barrel center entrance, helix α7 (green) and iii. catalytic loop L26. (a) The native Met549 occupies the route for the substrate achieves the catalytic. On the other hand, the single mutation Met549Ser stabilizes aromatic residues Tyr264 and Trp219; anion-π interaction between hydroxyl of Ser549 residue and the Phe551 residue is created. Interactions between the Tyr264 and Trp219 residues and the anion at the Ser549 mutated residue keep the site opened near ring C. (b) Stabilization of the Ser549/Phe551/Trp219/Tyr264 complex. The anion-π interaction is distanced by 2.7 Å, whereas the T-shaped π-π interaction is 4.3 Å closed.