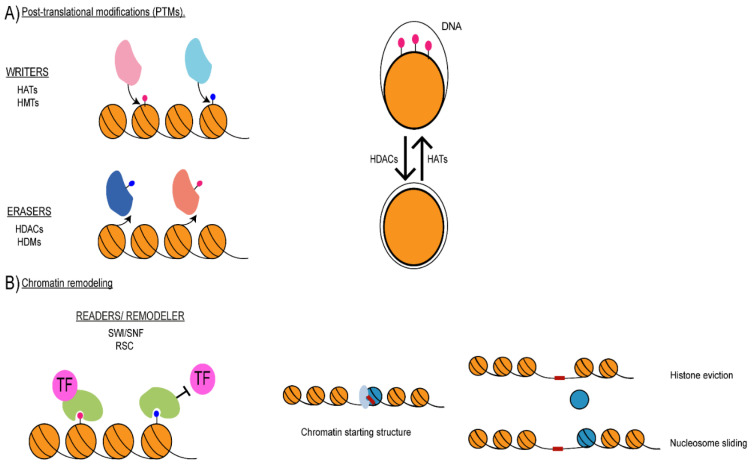
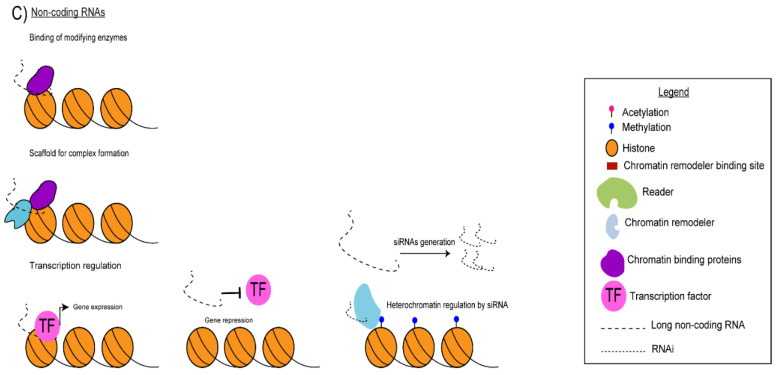
Figure 2.
Mechanisms of modulation in chromatin structures. (A) Left: schematics of chromatin writers and erasers: writers, such as HATs and HMTs, add epigenetic marks to histone proteins, while erasers, such as HDACs and HDMs, remove epigenetic marks from histone proteins. Right: schematic of how post-translation modifications, such as histone acetylation, can affect DNA-histone interactions. (B) Modes of action of readers and chromatin remodelers. Left: reader proteins, components of chromatin remodelling complex, bind modified histone tails recruiting or blocking transcription factors. Right: schematics of chromatin remodellers’ activity, including histone eviction and nucleosome sliding. (C) Mode of action of non-coding RNAs (ncRNAs). Top: ncRNAs can recruit modifiers to chromatin or act as a scaffold promoting the formation of protein complexes. Middle: ncRNAs can activate or repress transcription. ncRNAs can also be processed into siRNAs by the RNAi machinery. siRNAs can seed heterochromatin formation. HAT: histone acetyltransferase; HMT: histone methyltransferase; HDAC: histone deacetylase; HDM: histone demethylase; RSC: remodels the structure of chromatin.