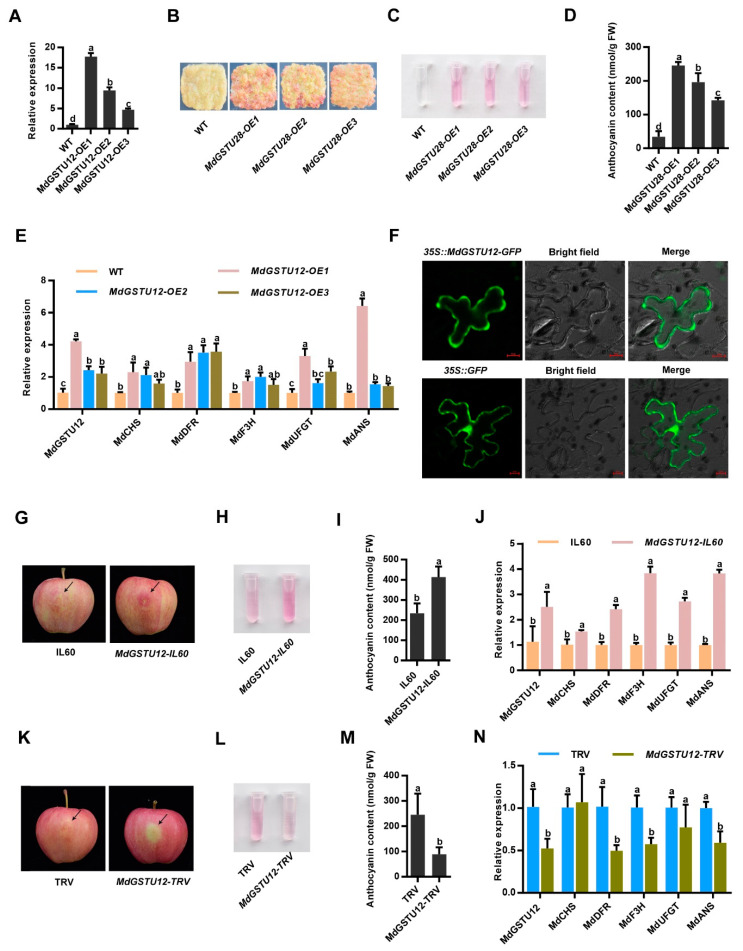
Figure 4.
MdGSTU12 is involved in the regulation of anthocyanin accumulation and subcellular localization of MdGSTU12 protein. (A) The expression of MdGSTU12 in the wild-type (WT) and MdGSTU12 transgenic apple calli by RT-qPCR assay. (B,C) Calli coloration of two-week-old WT, MdGSTU12-OE1, MdGSTU12-OE2, and MdGSTU12-OE3 transgenic calli. (D) Anthocyanin contents in the apple calli are shown in (B). (E) Expression analysis of MdGSTU12 and the anthocyanin biosynthesis-related genes MdCHS, MdDFR, MdF3H, MdUFGT, and MdANS by RT-qPCR assays in (B). (F) Subcellular localization of MdGSTU12 protein. 35S::GFP served as a control, scale bar = 10 µm, BF represents a bright field. (G) Coloration of apple fruit peels injected with plasmid mixtures (IL60: IL60-1 + IL60-2; MdGSTU12-IL60: IL60-1 + MdGSTU12-IL60-2). An empty IL60 vector was used as control. (H,J) Anthocyanin content (H,I) and relative expression levels of the anthocyanin biosynthesis-related genes (J) around the injection sites of the fruit peels shown in (G). (K) Coloration of apple fruit peels injected with a mixed solution of Agrobacterium cells (TRV: TRV1 + TRV2; MdGSTU12-TRV: TRV1 + MdGSTU12-TRV2). An empty TRV vector was used as a control. (L–N) Anthocyanin contents (L,M) and transcript levels of anthocyanin biosynthesis-related genes (N) around the injection sites of the fruit peels shown in (K). The 18s gene acted as the internal control. In A, D, E, I, J, M, N, the error bars indicate the standard deviation (SD) of three independent experiments, each of which included three technical replicates. Different lowercase letters indicate a significant difference at p < 0.05.