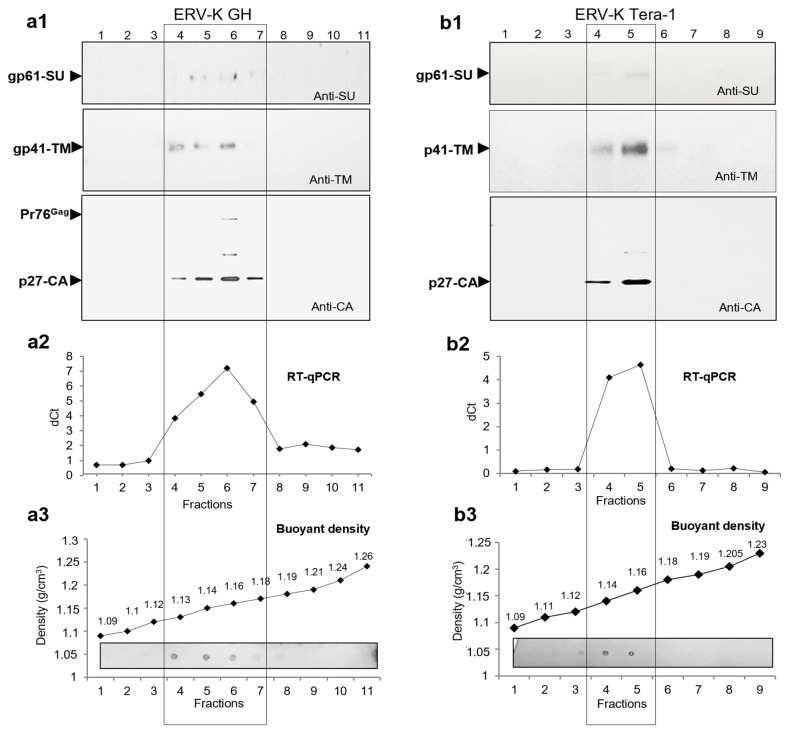
Figure 4.
Searching for diversification in HML.2 GH pool and analyses of SU, TM and vRNA in fractions of the gradient. (a1) analyses of HML.2 GH using extended 20–60% sucrose gradient. Viral SU, MA and CA proteins were detected by Western blot analyses; (a2) viral RNA in fractions of the gradient was analyzed by RT-qPCR. A minor hump seen on the cDNA curve F4 (1.13 g/cm3) most likely represented fragmented virus elements in association with “light” MVs, but not a separate population of assembled viruses; (a3) buoyant density of gradient fractions and total protein content in fractions using dot blot (lower panel) are shown; (b1) analyses of HML.2 Tera-1 were performed in a similar way, but using a standard 20–60% sucrose gradient. Signs of the virus’s diversification were not detected. A low amount of SU was shown in both preparations; (b2) viral RNA distribution along the gradient; (b3) buoyant density of gradient fractions and total protein content in fractions of the gradient estimated by a dot blot (low panel). Western blot analyses were performed using rabbit anti-SU serum (upper panel), mouse anti-TM Mab (middle panel) and rabbit anti-CA (low panel). Viral RNA content was examined by RT-qPCR. Delta threshold cycle (Ct) values were obtained by subtraction of RT-qPCR Ct values from Ct values did during qPCR without RT step. Virus-specific proteins are marked with arrow heads. Fractions with the highest protein and vRNA contents are in frame.