Abstract
COVID-19 pandemic poses a serious threat to human health; it has completely disrupted global stability, making vaccine development an important goal to achieve. Monoclonal antibodies play an important role in subunit vaccines strategies. In this work, nine murine MAbs against the RBD of the SARS-CoV-2 spike protein were obtained by hybridoma technology. Characterization of purified antibodies demonstrated that five of them have affinities in the order of 108 L/mol. Six MAbs showed specific recognition of different recombinant RBD-S antigens in solution. Studies of the additivity index of anti-RBD antibodies, by using a novel procedure to determine the additivity cut point, showed recognition of at least five different epitopes. The MAbs CBSSRBD-S.11 and CBSSRBD-S.8 revealed significant neutralizing capacity against SARS-CoV-2 in an ACE2-RBD binding inhibition assay (IC50 = 85.5pM and IC50 = 122.7pM, respectively) and in a virus neutralizing test with intact SARS-CoV-2 (VN50 = 0.552 nM and VN50 = 4.854 nM, respectively) when D614G strain was used to infect Vero cells. Also CBSSRBD-S.11 neutralized the SARS-CoV-2 strains Alpha and Beta: VN50 = 0.707 nM and VN50 = 0.132 nM, respectively. The high affinity CBSSRBD-S.8 and CBSSRBD-S.7 recognized different epitopes, so they are suitable for the development of a sandwich ELISA to quantitate RBD-S recombinant antigens in biomanufacturing processes, as well as in pharmacokinetic studies in clinical and preclinical trials.
Keywords: SARS-CoV-2, COVID-19, Receptor binding domain, Neutralizing monoclonal antibodies, Additivity index
1. Introduction
The coronavirus induced disease (COVID-19) was unknown until outbreak in December 2019 when the first patients were detected in Wuhan, China, and the studies identified a new coronavirus as etiological agent: the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Gorbalenya et al., 2020; Zhou et al., 2020). A few months after its epidemic outbreak COVID-19 became a pandemic that represents a serious threat to human health and has changed entire worldwide dynamics affecting not only public health but education, economy and so many other spheres (UN News, 2020; WHO, 2020). Globally, there have been 3,937,437 deaths from COVID-19, reported to the WHO as of June 30, 2021 (WHO, 2021b). In addition, novel variants of SARS-CoV-2 have emerged during human to human transmission and its long-term consequences are still under study (GOV.UK, 2021a, GOV.UK, 2021b; Valdés et al., 2021; Valdes-Balbin et al., 2021; Zhang et al., 2021).
Coronaviridae is a broad family of viruses including SARS-CoV-2 that it is very similar to SARS-CoV but ostensibly more contagious due to a higher affinity to the receptor angiotensin-converting enzyme 2 (ACE-2) which is expressed on numerous cells, including lung, heart, kidney, and intestine cells. Its host-cell infection mechanism involves the subunit 1 (S1) located in the N-terminal of the spike protein (S) that recognizes and binds ACE-2 through the receptor-binding domain (RBD) (Chen et al., 2020; Gorbalenya et al., 2020; Kuba et al., 2005; Li et al., 2003; Yan et al., 2021; Yan et al., 2020).
The availability of safe and effective vaccines has shown to represent a radical change in the face of the COVID-19 pandemic (WHO, 2021c). Subunit vaccines stand out among the different development strategies due to their advantages of safety, stability and more feasible storage conditions compared to others, 30% of vaccines in clinical development are been deployed in subunit platform (WHO, 2021d). Several vaccines strategies against SARS-CoV-2 target the S protein because it induces a high immune response in humans and specifically the receptor-binding domain of S protein (RBD—S) induces the highest titers of neutralizing antibodies (He et al., 2004; Kyriakidis et al., 2021; Limonta-Fernández et al., 2021; Qi et al., 2020; Valdés et al., 2021; Valdes-Balbin et al., 2021; Zhu et al., 2013).
MAbs are displaying a crucial role in the investigations related to COVID-19, which are focused in effective therapies, vaccines design and SARS-CoV-2 detailed characterization (Taylor et al., 2021; Valdes-Balbin et al., 2021). Many neutralizing MAbs against SARS-CoV-2 have been reported, among which the recombinant or native human MAbs stand out (Andreano et al., 2020; Bertoglio et al., 2021; Wang et al., 2020; Yuan et al., 2021), but also those produced in plants such as Nicotiana benthamiana (Rattanapisit et al., 2020) or isolated from immunized rabbits to be produced in VeroE6 cells (Makdasi et al., 2021). Other investigations report the obtainment of mouse MAbs by hybridoma technology (Chapman et al., 2021; Guo et al., 2021) and its subsequent evaluation for application in molecular pathology as humanized MAbs (Guo et al., 2021).
Several anti SARS-CoV-2 MAbs entered clinical trials during the second half of 2020 (Marovich et al., 2020). Until date, six neutralizing MAbs obtained an Emergency Use Authorization by the regulatory agencies of United States and South Korea, and others are being evaluated in clinical trials. MAbs authorized or in development are directed to the RBD, all of them are fully human and the majority was obtained from SARS-CoV-2-immune donors (Corti et al., 2021).
Furthermore, murine MAbs are being applied as biological reagents offering analytical support determination of SARS-CoV-2 spike antigens by design of various immunodetection systems required in biomanufacturing, diagnostic and virus neutralization tests (Kim et al., 2021; Lee et al., 2021; Perera et al., 2020; Tan et al., 2020).
This work describes the obtainment of mouse MAbs with specific recognition of the recombinant RBD-S protein of SARS-CoV-2 suitable for analytical methods required in the research-development and production of subunit vaccines as well as in clinical trials. MAbs were characterized with respect to important features they should have in order to be used as reagents such as: affinity, specificity against different recombinant RBD antigens, epitope specificity among each MAb pairs and potential neutralizing activity. The neutralizing murine MAbs could be humanized in order to test their therapeutic potential.
2. Materials and methods
2.1. Antigens
Different recombinant antigens (Table 1 ) corresponding to fragments of the RBD-S of SARS CoV-2. RBDr1, RBDr4 were supplied by the Center for Molecular Immunology, RBDr2 (Limonta-Fernández et al., 2021) and RBDr3 by the Center for Genetic Engineering and Biotechnology and RBDr5 (Valdes-Balbin et al., 2021) by the Finlay Institute of Vaccines, all located in Havana, Cuba. RBDs contain different C-terminal or N-terminal fusion tags.
Table 1.
Recombinant RBD antigens used in this research.
| Antigens | Host cell | S protein fragment from | Tags |
|---|---|---|---|
| RBDr1 | HEK293T | Arg328 – Leu533 | 6xHis |
| RBDr2 | Pichia Pastoris | Asn331 – Ser 529 | 6xHis |
| RBDr3 | HEK293T | Asn331 – Ser 529 | 6xHis |
| RBDr4 | HEK293T | Arg328 – Leu533 | human IgG- Fc |
| RBDr5 | CHO | Arg319 – Phe541 | 6xHis |
A sequence coding for residues of the Spike protein of SARS-CoV-2 was based on Wuhan-Hu-1 strain sequence (NCBI Acc. No. YP_009724390).
2.2. Immunization schedule
BALB/c female mice with a body weight between 20 and 25 g were supplied by CENPALAB, Cuba. Before starting the immunization schedule, blood samples were collected from each mouse to be used as pre-immunized control serum. Mice received three subcutaneous immunizations with intervals of 15 days. The first dose was 100 μg of RBDr1 emulsified in Freund's complete adjuvant. The second and third immunizations were administered with 50 μg of antigen in incomplete Freund's adjuvant. Seven days after the last injection, blood was obtained for serum titration of specific antibodies by ELISA. Three days before the splenectomy, the mouse with highest serum titer was inoculated intraperitoneally with a 50 μg booster dose of RBDr1 dissolved in sterile phosphate buffered saline (PBS, 0.137 M NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4). The experimental protocols were approved by the Ethical Committee on Animal Experimentation of the Center for Genetic Engineering and Biotechnology (CIGB, Havana, Cuba).
2.3. Indirect ELISA for evaluating anti-RBD-S antibodies
Costar® 3590 high binding plates were coated with 100 μL/well of 10 μg/mL of RBDr1 diluted in 0.01 M carbonate-bicarbonate buffer solution, pH 9.6 and incubated 2 h at 37 °C. Once washed three times with PBS-0.05% Tween 20 (PBST) the plates were blocked with a 3% (w/v) nonfat dried milk solution in PBS during 1 h at 37 °C and next the wells were washed once with PBST. Subsequently, the samples: eleven serial dilutions, from 1/500 to 1/51200, of sera from immunized mice (diluted in PBS with 1% (w/v) nonfat dried milk) and undiluted cell culture supernatant, were applied 100 μL/well. After samples incubation 2 h at 37 °C there were made three PBST washes and there were added 100 μL/well of a horseradish peroxidase (HRP)-labeled sheep anti-mouse IgG antibodies (Sigma, USA), diluted 1:10000 in PBS with 1% (w/v) nonfat dried milk, 1 h at 37 °C. Finally, plates were washed three more times and incubated in the dark with 100 μL/well orto-phenylenediamine (0.5 mg/mL, dissolved in 0.0243 M citric acid, 0.0514 M disodium hydrogen phosphate and 0.015% (w/v) hydrogen peroxide) for 20 min at RT. The reaction was stopped with 100 μL/well 2 M sulfuric acid and the absorbance at 492 nm (A492nm) was measured by the ELISA plate reader (Labsystems Multiskan® Plus, Finland).
2.4. Generation of anti-RBD-S MAbs
Spleen cells from higher titer immunized mouse were fused with exponentially growing mouse myeloma cells line P3X63Ag8.653 three days after booster injection. A mixture of 100 × 106 spleen cells and 10 × 106 myeloma cells was fused using 50% (w/v) polyethylene glycol solution (Sigma, Hybri-Max) according to a published modification of the Köhler and Milstein protocol (Galfrè and Milstein, 1981; Köhler and Milstein, 1975). The cells were cultured in RPMI 1640 (Gibco, USA) supplemented with 20% fetal bovine serum (FBS) (Capricorn Scientific, origin Australia), hipoxantine, aminopterine and thymidine (HAT) selection medium additive (Sigma Hybri-Max, USA) and were distributed in 96-well culture plates. The plates were incubated at 37 °C in a 5% CO2 atmosphere. The culture supernatants of the hybrid cell colonies were tested for anti-RBD-S antibodies secretion by the specific ELISA described above. Hybridomas from ELISA positive wells were collected and repetitively cloned by limiting dilution method at a rate of one cell/well.
2.5. Determination of MAbs isotypes
Identification of MAbs class and subclass was carried out using a mouse monoclonal antibody isotyping kit (Sigma, Germany) according to the manufacturer's recommendations. Plates were coated with 100 μL/well of 10 μg/mL of RBDr1 diluted in 0.01 M carbonate-bicarbonate buffer solution, pH 9.6 and incubated 2 h at 37 °C.
2.6. Production and purification of MAbs
The selected hybridoma cell lines were washed in RPMI-1640 medium (Gibco, USA), centrifuged at 180 ×g for 10 min at RT and resuspended in RPMI-1640 medium, until a concentration of 2 × 106 cells/mL. The cell suspension was injected intraperitoneally to BALB/c mice, 1 mL per mouse. Ten days before the injection, the mice peritoneum was stimulated with paraffinic mineral oil. Seven days after, the ascites fluid was extracted by puncture of the peritoneum and clarified by centrifugation at 1125 ×g for 30 min at RT.
The ascites fluid was filtered through 0.45 μm glass wool, precipitated with 50% (w/v) of ammonium sulfate and centrifuged 30 min at 6000 ×g. The pellet was resuspended in 1.5 M glycine, 3 M NaCl, pH 8.9, and loaded onto an nProtein A Sepharose Fast Flow matrix (Cytiva, USA). Elution was carried out with 0.1 M citrate buffer, pH 6.0. The chromatographic profile was checked by measuring the absorbance at 280 nm. The eluate was neutralized with 2 M Tris-HCl and dialyzed in 20 mM Tris-HCl, 150 mM NaCl, pH 7.0, during 16 h, and then filtered through 0.2 μm membrane. A final concentration of 0.02% (v/v) of thimerosal was added as preservative. Purified samples were evaluated by densitometric analysis of SDS-PAGE 12.5%.
2.7. Affinity constants of the anti- RBD-S monoclonal antibodies
To measure the affinity constant of eight MAbs generated against the recombinant SARS-CoV-2 Spike Protein receptor binding domain (RBDr1) the methodology of Beatty et al. was followed(Beatty et al., 1987), by the indirect antibody capture ELISA used for hybridoma screening described above, with modifications. Eight plates were divided into four sections of two rows each one and coated from bottom to top with concentrations of 1.25, 2.5, 5 and 10 μg/mL of RBDr1. Each antibody was tested on a separate plate by performing 1:4 serial dilutions from the third column to the 12th, the concentrations were between 10 μg/mL and 38 pg/mL. First and second columns of all plates were reserved as blanks of the assay. The affinity constant (Kaff) was calculated by the formula:
where n represents the ratio between the highest and the lowest antigen concentration for each of the six possible comparisons between the four antigen concentrations used. In a comparison between two antigen concentrations, [Ab]1 represents the molar antibody concentration calculated for A-50 (half of maximum A492nm), corresponding to the lower antigen concentration. [Ab]2 represents the molar antibody concentration calculated for A-50 measured at 492 nm, corresponding to the highest antigen concentration. The calculation of [Ab]1 and [Ab]2 was carried out by interpolating the value of A-50 in the curve of A492nm vs. antibody concentration, fitting the curve to a five-parameters logistic regression by GraphPad Prism version 8.0.2 for Windows, GraphPad Software, San Diego, California USA.
The Kaff value in L/mol for each antibody represents the mean ± the standard deviation (SD) of the six calculated Kaff values.
2.8. Evaluation of the specificity of the MAbs against the RBDr2 protein
In this procedure, the indirect ELISA described above was applied, with the difference that it includes a step of inhibition in solution of the MAbs with the recombinant antigens RBDr1 and RBDr2 for 1 h at RT. The antigen was added in each inhibition mixture in molar concentrations greater than 100 times the concentration of the MAbs. Inhibition was calculated as follows:
The A492nm values were the mean of triplicate measurements.
2.9. Additivity test by ELISA
The additivity test described by Friguet and colleagues (with adjustments in the cut point calculation procedure) was applied in order to evaluate whether two MAbs recognize different antigenic sites (Friguet et al., 1983). This test was performed using the previously described indirect ELISA with small modifications. The plates were coated with 100 μL/well of 0.7 μg/mL of RBDr1. Each pair of MAbs was added at saturating concentration for this coating (10 μg/mL). The additivity index (AI) of a pair of antibodies was defined as follows:
where A1, A2 and A1 +2 are the A492nm reached in the ELISA with the first MAb alone, the second MAb alone, and the two MAbs together, respectively. If the two antibodies randomly bind to the same site then A1 +2 must be equal to the mean value of A1 and A2 and the additivity index should theoretically equal to 0%. Conversely, if the two antibodies bind independently to different sites, A1 +2 must be the sum of A1 and A2 and AI should be theoretically equal to 100%. To establish an objective cut point of AI values that indicate that two antibodies recognize different antigenic sites, the AI of the same antibody (AIzero) was calculated. To this aim, it was established that A1 and A2 represent the experimental absorbance values of the same antibody at two different wells on the ELISA plate and A1 +2 represents the experimental absorbance value of the same antibody at double saturating concentration. The cut point (CP) was defined as:
where the AIzero mean and the SD of the seven MAbs tested are used, t0.05,df is the one-tail critical value determined from t-distribution corresponding to a 5% false-additivity rate and “df” are the degrees of freedom that depend on the numbers of MAbs analyzed.
2.10. Sandwich ELISA for RBDr2 antigen quantification
Costar® 3590 high binding polystyrene 96-well microtiter plates were coated with 100 μL/well of MAb CBSSRBD-S.8, at 5 μg/mL in 0.1 M carbonate-bicarbonate buffer, pH 9.6, for two hours at 37 °C. The plates were then washed tree times with PBST. These washing conditions were applied in all steps of the ELISA, except after blocking. Then 380 μL/well of blocking solution (3% (w/v) nonfat dried milk powder in PBS) was added to the plates and incubated for 1 h at 37 °C. After one washing, 100 μL/well of RBDr2 as antigen were added in concentrations from 0.12 ng/mL to 30 ng/mL diluted in 2% (w/v) nonfat dried milk powder in PBS. The plate was incubated 1 h at 37 °C and next a wash step was performed. The MAb CBSSRBD-S.1 and MAb CBSSRBD-S.7 were previously conjugated to horseradish peroxidase by the method proposed by (Wilson and Nakane, 1978). The CBSSRBD-S.1-HRP and CBSSRBD-S.7-HRP were diluted 1: 1600 or 1: 8000, respectively in PBS with 1% (w/v) nonfat dried milk powder. This step was performed using 100 μL/well of each respective (HRP)-labeled MAb and incubated for 1 h at 37 °C. Finally, plates were washed and incubated in the dark with 100 μL/well orto-phenylenediamine (0.5 mg/mL, dissolved in 0.0243 M citric acid, 0.0514 M disodium hydrogen phosphate and 0.015% (w/v) hydrogen peroxide) for 20 min at RT. The reaction was stopped with 100 μL/well 2 M sulfuric acid and the A492nm was measured by the ELISA plate reader. Nonfat dried milk powder in PBS solution at 2% (w/v) was used as assay blank.
The assay was repeated during six non-consecutive days by two analysts (three assays per analyst). The interassay precision and accuracy of the back-calculated values for each standard concentration were used to determine the limits of quantification. The evaluation of precision was performed using the percentage of coefficient of variation (CV), calculated as CV = (SD/mean concentration) × 100. The evaluation of accuracy was performed using the relative error, calculated as error (%) = (calculated mean concentration - nominal concentration) × 100/nominal concentration. The lower limit of quantification was defined as the minimum value of the standard curve whose precision and accuracy was ≤20%. Correspondingly, the maximum value of the standard curve whose precision and accuracy was ≤15% was defined as the upper limit of quantification. This ELISA was also used for the quantification of the four other recombinant RBD-S antigens (Table 1). The ELISA standard curves were fitted to five-parameter logistic regression by GraphPad Prism software version 8.0.2 for Windows.
2.11. hFc-ACE-2 and RBDr4-HRP binding inhibition by the monoclonal antibodies
Costar® 3590 high binding polystyrene 96-well microtiter plates were coated with 50 μL/well of hFc-ACE-2 recombinant protein expressed in HEK293T (supplied by the Center for Molecular Immunology, Cuba), at 5 μg/mL in PBS, pH 7.4, during three hours at 37 °C. After one washing with 0.1% (v/v) of Tween-20 in distilled water, the plates were blocked with 2% (w/v) nonfat dried milk powder in PBS for 1 h at 37 °C. The MAbs in concentrations of 5, 10, 50, 100, 500 and 1000 ng/mL as well as the test controls were pre-incubated for 1 h at 37 °C with RBDr4-HRP, conjugated according to Wilson and Nakane protocol (Wilson and Nakane, 1978) diluted 1:100000 with 0,2% (w/v) skim milk powder in PBS. A high titer SARS-CoV-2 convalescent serum was used as positive control. Human AB Serum (Cat. No.H4522, Sigma, USA) was employed as negative control.
After pre-incubation period, 50 μL of sample-conjugate mixtures were added to hFc-ACE-2 blocked plate and incubated during 1 h at 37 °C. The plate was washed four times with 0.1% (v/v) of Tween 20 in distilled water and incubated in the dark for 10 min at RT with 50 μL/well of 3,3′,5,5′-tetramethylbenzidine at 10 μg/mL, dissolved in phosphate-citrate buffer (0.2 M phosphate, 0.1 M citrate, pH 5.0) and 0.006% (v/v) hydrogen peroxide. The reaction was stopped with 50 μL of 2 M sulfuric acid. The absorbance was measured at 450 nm, using the ELISA plate reader.
Inhibition of the MAbs to the binding of RBDr4-HRP and hFc-ACE-2 was calculated as follows:
where A450 Inh means A450nm of RBDr4-HRP preincubated with the MAb and A450 Max means the A450nm of the non-inhibited RBDr4-HRP. The strength of inhibition of the MAbs was estimated by the half-maximum inhibitory concentration (IC50). The IC50 was calculated by fitting the data to a five-parameter logistic regression with GraphPad Prism software version 8.0.2 for Windows.
2.12. Microneutralization of live SARS-CoV-2 virus in Vero E6 cell line
The neutralization antibody capability of the MAbs CBSSRBD-S.8 and CBSSRBD-S.11 were evaluated by a traditional virus microneutralization assay according to (Limonta-Fernández et al., 2021) using the SARS-CoV-2 strains D614G, and the variants of concern Alpha (lineage B.1.1.7), Beta (lineage B.1.351) and Delta (lineage B.1.617.2). The MAbs were tested against the D614G strain in a concentration range of 0.109 nM to 41.7 nM. The viral neutralization percentage (VN) of MAbs was measured as A540nm sample x 100/ A540nm control. A540nm sample represents a directly proportional signal to MAbs neutralizing activity and A540nm control represents a signal of intact cells (no virus added). The viral neutralizing at 50% of maximum VN (VN50) was calculated by fitting the curve to a five-parameter logistic regression using GraphPad Prism software version 8.0.2 for Windows.
3. Results and discussion
3.1. Mice serum titration
The highest antibody titers reached in serum from immunized mice was in order of 1:8900 at day 37 after the first immunization. These results were comparable with those obtained by Zakhartchouk et al. in a different SARS-CoV spike protein immunizations schedule (Zakhartchouk et al., 2007), furthermore other authors reported a sub-optimal immunogenicity of RBD from SARS-CoV-2 (Mandolesi et al., 2020; Tan et al., 2021). Therefore, the best tittered mouse was splenectomized and its splenocytes properly processed for the fusion with the myeloma cells.
3.2. Generation of stably hybridoma cell lines and purification of MAbs
Hybridomas obtained after fusion, were seeded in seventeen 96 wells plates. A total of 1632 wells were seeded. Ten days after the fusion 1510 wells were observed with hybrid cells growth that survived to the HAT supplemented selection medium, which represents a 92.5% of fusion efficiency.
The ELISA screening of the cell culture supernatant allowed detecting twenty two wells as putative positive for secretion of specific MAbs against RBD—S, but only eight stable hybridomas cells line were obtained. Each one of them was three times cloned by limiting dilution method to ensure that cells that produce the antibody of interest were single-cell cloned and, consequently, the secretion of this antibody can be stably maintained. The hybridomas clones and its respective secreted MAbs were named (Table 2 ) and isotyped as IgG1. These results confirmed that the immunization schedule, as well as the concentration of the antigen used was suitable for obtaining high fusion efficiency and to develop a successful generation of hybridomas.
Table 2.
Affinity constants of MAbs against RBDr1.
| Monoclonal antibodies | Kaff (L/mol) |
|---|---|
| CBSSRBD-S.1 | 3,34 ± 2,76 × 107 |
| CBSSRBD-S.2 | 8,10 ± 4,92 × 106 |
| CBSSRBD-S.3 | 4,40 ± 1,60 × 106 |
| CBSSRBD-S.4 | 1,15 ± 0,39 × 108 |
| CBSSRBD-S.5 | 2,77 ± 2,18 × 107 |
| CBSSRBD-S.6 | 1,04 ± 0,60 × 108 |
| CBSSRBD-S.7 | 1,59 ± 0,26 × 108 |
| CBSSRBD-S.8 | 1,57 ± 0,60 × 108 |
Values for each antibody represent the mean ± SD of the six calculated Kaff.
The selected hybridoma cell lines were inoculated in BALB/c mice for ascites production. Ascites tumors were obtained in 100% of inoculated animals. MAbs purification from ascites was carried out by Protein A Sepharose affinity chromatography. Final yields obtained were between 1.0 and 2.5 mg of purified MAbs per mL of ascitic fluid. The purity of MAbs was greater than 99%.
3.3. Affinity constants of the anti-RBD MAbs
Four of the eight antibodies evaluated have affinity constant in the order of 108L/mol. The MAbs CBSSRBD-S.7 and CBSSRBD-S.8 antibodies showed the highest affinity with very similar values (Table 2).
3.4. Evaluation of the specificity of the MAbs against the RBDr2
The specificity of the MAbs generated against the RBDr1 antigen was evaluated by an inhibition assay with the RBDr1 and RBDr2 antigens. Even when the MAbs were generated against the RBDr1 protein, their specificity against this antigen was evaluated as a control for the functionality of the assay.
As expected, MAbs were highly inhibited by the antigen against which they were generated (RBDr1), except CBSSRBD-S.2 and CBSSRBD-S.3, which only inhibited 78.79% and 76.79%, respectively. This result was consistent with the fact that they were the lowest affinity antibodies (Table 2). Furthermore, both were the only MAbs that recognized RBDr1 under denaturing Western blot conditions (data not shown). This could suggest that these antibodies target a linear epitope partially occluded in a fully folded RBD molecule. The inhibition percentages with RBDr2 were equally high evidencing specific antigen-antibody recognition in solution, except with the MAbs CBSSRBD-S.2 and the CBSSRBD-S.3 (Table 3 ). In this case, the recognition site for these MAbs could be absent in RBDr2, the shortest molecule of the antigens tested. This idea could be supported by the fact of no signal detection in Western blot (data not shown). These highly specific antibodies could be useful for the development of assays that require the recognition of both recombinant and natural antigens.
Table 3.
Assessment of the specificity of MAbs against recombinant antigens RBDr1 and RBDr2.
| Sample | A492nm (non-inhibited samples) | Inhibition (%) |
|||
|---|---|---|---|---|---|
| RBDr1 | RBDr2 | RBDr1 | RBDr2 | ||
| CBSSRBD-S.1 | 0.815 | 0.002 | 0.004 | 99.75 | 99.51 |
| CBSSRBD-S.2 | 0.264 | 0.056 | 0.278 | 78.79 | −5.43 |
| CBSSRBD-S.3 | 0.457 | 0.106 | 0.428 | 76.79 | 6.20 |
| CBSSRBD-S.4 | 0.975 | 0.004 | 0.007 | 99.56 | 99.25 |
| CBSSRBD-S.5 | 0.795 | 0.001 | 0.007 | 99.83 | 99.16 |
| CBSSRBD-S.6 | 0.916 | 0.004 | 0.002 | 99.60 | 99.75 |
| CBSSRBD-S.7 | 0.759 | 0.016 | 0.126 | 97.89 | 83.44 |
| CBSSRBD-S.8 | 0.604 | 0.002 | 0.001 | 99.61 | 99.89 |
Inhibition (%) = [1- A492nm (inhibited samples)/A492nm (non-inhibited samples)] × 100. The absorbance values are the mean of triplicate measurements with SD within 20% of the means.
3.5. ELISA additivity test
The epitope competition between two antibodies was analyzed by an additivity ELISA. This assay requires the antigen to be saturated for each antibody tested. In order to ensure accuracy in determining the AI, it was established that coating the plate with 0.7 μg/mL of RBDr1 and adding the MAbs in saturating concentrations (10 μg/mL) guaranteed A492nm values between 0.400 and 0.500 for each antibody alone on the ELISA plate. The AI values of each pair of MAbs are shown in Table 4 . The AI data arranged on the diagonal and indicated in bold correspond to the AIzero. An arbitrary cut point is generally used in this kind of analysis e.g. (Haggarty et al., 1986; Liu et al., 2010; Zai et al., 2018) but this could affect the classification of antibodies in terms of the recognition of antigenic sites. Therefore, this work establishes a procedure to determine the cut point taking into account the distribution of random errors of the assay signal (A492nm). The use of the AIzero (additivity index of the same antibody) is the closest consideration to the AI obtained between two antibodies that compete for the same epitope. The cut point for AI in this trial was 41%, calculated from the formula described above and using t 0.05, 6 df = 1.943, SD = 16.06% and the mean of the IAzero = 9.79%.
Table 4.
Additivity index (%) of the seven MAbs.
| MAbs | CBSSRBD-S.1 | CBSSRBD-S.2 | CBSSRBD-S.3 | CBSSRBD-S.4 | CBSSRBD-S.6 | CBSSRBD-S.7 | CBSSRBD-S.8 |
|---|---|---|---|---|---|---|---|
| CBSSRBD-S.1 | −8.05 | 88.27 | 47.15 | 13.90 | 4.46 | −16.09 | 73.61 |
| CBSSRBD-S.2 | 34.93 | 62.73 | 56.18 | 56.32 | 75.06 | 100.53 | |
| CBSSRBD-S.3 | 18.83 | 61.04 | 57.09 | 59.86 | 75.09 | ||
| CBSSRBD-S.4 | −3.13 | 14.14 | −1.35 | 67.66 | |||
| CBSSRBD-S.6 | 1.83 | 2.59 | 56.16 | ||||
| CBSSRBD-S.7 | 0.34 | 78.90 | |||||
| CBSSRBD-S.8 | 23.80 |
Data represent the AI calculated from triplicate absorbance values at 492 nm. The coefficient of variation of the absorbance values was less than 20%. The AI data arranged on the diagonal and highlighted in bold type, correspond to the AIzero. The mean of the AIzero was 9.79% and the SD was 16.06%. The cut point calculated, at 95% confidence, was AI≥41% (see formula defined above, for a t0.05, 6df value of 1.943). If the two antibodies bind randomly at the same site the AI will be ˂41%. If the two antibodies bind independently at distinct sites AI will be ≥41%.
The AI of all possible combinations of the CBSSRBD-S.1, CBSSRBD-S.4, CBSSRBD-S.6 and MAbs CBSSRBD-S.7 (Table 4) showed that the binding of each pair of MAbs was not additive suggesting that these MAbs recognize the same epitope or very close epitopes, such that the binding to the antigen of one MAb causes a steric hindrance to the binding of the other Mab, or that the binding of one MAb leads to a conformational change in the antigen preventing the binding of the second MAb. In contrast, the AIs of the MAbs CBSSRBD-S.2, CBSSRBD-S.3 and CBSSRBD-S.8 showed they are additive among them and with the MAbs CBSSRBD-S.1, CBSSRBD-S.4, CBSSRBD-S.6 and CBSSRBD-S.7. This result suggested that the generated MAbs recognize at least four different epitopes of RBDr1. A first epitope is recognized by the MAbs CBSSRBD-S.1, CBSSRBD-S.4, CBSSRBD-S.6 and CBSSRBD-S.7, a second epitope by MAb CBSSRBD-S.2, a third epitope by the MAb CBSSRBD-S.3 and a fourth epitope by the MAb CBSSRBD-S.8. The AI data presented, in correlation with their affinity values (Kaff), allow the optimal selection of a pair of MAbs for the development of a sandwich type immunoassay for the quantification of recombinant antigens of RBD-S in their production processes and/or for pharmacokinetic studies of recombinant antigens in pre-clinical and clinical trials.
3.6. Sandwich ELISA for the quantification of RBD antigen
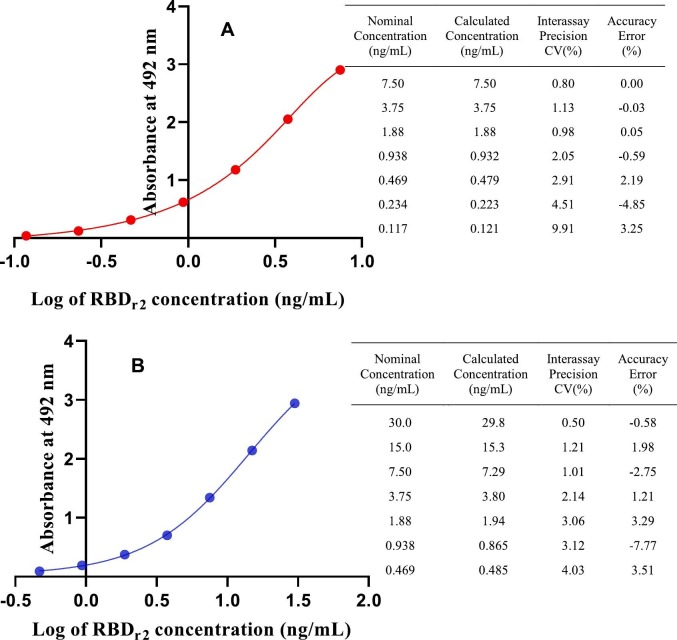
Since the MAbs CBSSRBD-S.8 and CBSSRBD-S.7 are the antibodies with the highest affinity (Table 2) and recognize different epitopes (Table 4), they were selected to develop a sandwich ELISA that aimed to be used for quantification of RBD-S recombinant antigens in their production processes, as well as in the pharmacokinetic studies of RBD based vaccine candidates against SARS-CoV-2. As other authors have recommended (DeSilva et al., 2003) for standard curve fitting in immunoassays, five-parameter logistic regression was the best fit obtained for this RBDr2 standard curve (Fig. 1A). All points on the antigen curve were back-calculated with interassay precision and accuracy less than 9.91% and 4.85%, respectively. Standard curves are considered suitable at accuracy and precision ≤15% for each point, except at the lower limit of quantification, which could be ≤20% (DeSilva et al., 2003). The range of quantification of standard curve was 0.121 ng/mL to 7.50 ng/mL. Due to the nature or format of many immunoassays, the quantification range of the standard curve can be very narrow, sometimes less than an order of magnitude (DeSilva et al., 2003). The lower limit of quantification obtained with this combination of MAbs was 0.121 ng/mL. If it were possible to quantify this antigen at the minimum required dilution recommended for immunoassays, between 1/20 and 1/100 (Mire-Sluis et al., 2004) then this ELISA could be suitable for the detection of RBDr2 in concentrations between 2.42 and 12.1 ng/mL of cell culture supernatant or serum.
Fig. 1.
Five-parameter logistic fit for the sandwich ELISA standard curves. These were obtained by coating the plate with the MAb CBSSRBDS-8 and using the MAb CBSSRBD-S.7-HRP (A) or the MAb CBSSRBD-S.1-HRP (B) as conjugate. RBDr2 was used as standard for both curves. The adjacent tables show the respective coefficients of variation (CV) and mean error of the six ELISAs performed with each pair of antibodies as a measure of the precision and accuracy of the back-calculated concentration at each point.
In order to broaden the quantification range of RBDr2 the MAb CBSSRBD-S.1 was conjugated to horseradish peroxidase. The MAb CBSSRBD-S.1 recognizes a different epitope than the CBSSRBD-S.8 (Table 4) and has lower affinity than the CBSSRBD-S.7 (Table 2). With the new combination of MAbs (Fig. 1B), the five-parameter logistic regression was also the best fitted standard curve. All points on the antigen curve were back-calculated with interassay precision and accuracy of less than 9.05% and 15.92%, respectively. This new pair also allows a sensitive antigen determination, but with a wider quantification range for standard curve (0.272 ng/mL to 29.8 ng/mL). Taking into account the same minimum required dilution considerations with this antibody combination, this ELISA would be used for the detection of RBDr2 in concentrations between 5.44 and 27.2 ng/mL of cell culture supernatant or serum.
The CBSSRBD-S.8 and CBSSRBD-S.7-HRP sandwich also recognize four other antigen variants (Table 5 ). Given that RBDr2 is the shortest and most glycosylated variant, this result was predictable.
Table 5.
Quantification of the RBD-S recombinant antigen variants, evaluated with an ELISA sandwich combining the MAbs CBSSRBD-S.8 and CBSSRBD-S.7-HRP.
| Antigens | Quantification range (ng/mL) | Error (%) |
|---|---|---|
| RBDr1 | 0.460–30.0 | ≤ 0.75 |
| RBDr2 | 0.121–7.50 | ≤ 3.25 |
| RBDr3 | 0.460–30.0 | ≤ 5.81 |
| RBDr4 | 1.88–60.0 | ≤ 3.56 |
| RBDr5 | 0.030–19.5 | ≤ 9.95 |
The range of quantification was based on the lower and upper limits of quantification, calculated from three independent trials. The error (%) represents the maximum error obtained between all points of the curve, as a measure of the accuracy of the back-calculated concentration by a five parameters logistic regression.
3.7. RBDr4-HRP and hFc-ACE-2 binding inhibition by the anti-RBD monoclonal antibodies
A SARS-CoV-2 surrogate virus neutralization test (sVNT) that detects neutralizing antibodies targeting to RBD-S was performed. RBDr4 was selected to be conjugated to HRP considering that is human-Fc tagged and its large tag could avoid a possible interference of the enzyme conjugation in the RBD binding to the ACE-2 receptor. In addition, RBDr4 is a human recombinant protein with a folding and glycosylation profile similar to the viral protein.
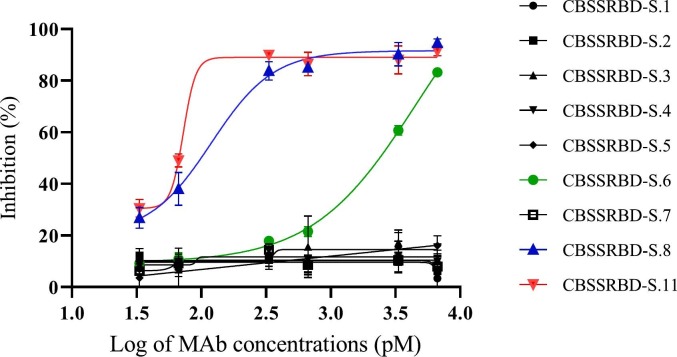
Two of the above analyzed MAbs strongly inhibited the binding of RBDr4-HRP and hFc-ACE-2 in the sVNT (Fig. 2 ). The IC50 calculated for the CBSSRBD-S.6 and CBSSRBD-S.8 MAbs was 2216.7pM and 122.7pM, respectively. These two MAbs have similar affinity constants (Table 2) and recognize different epitopes (Table 4). The greater binding strength of CBSSRBD-S.8 to RBDr4-HRP would be explained if this MAb recognizes an epitope more involved to the binding between the antigen and its receptor (RBDr4-HRP and hFc-ACE-2). On the other hand, even when the MAbs CBSSRBD-S.6, CBSSRBD-S.1, CBSSRBD-S.4 and CBSSRBD-S.7 have non-additive AI values, suggesting very close epitopes, only the CBSSRBD-S.6 inhibits the binding of RBD to ACE-2. The MAbs CBSSRBD-S.4 and CBSSRBD-S.7 that have similar affinities to CBSSRBD-S.6 could bind to epitopes not involved in ligand-receptor interaction. In the case of CBSSRBD-S.1, its affinity one order of magnitude less could cause non-appreciable neutralizing effect in this test, even if it recognized the same epitope as CBSSRBD-S.6. Recently, a new MAb (CBSSRBD-S.11), obtained in our lab from a longer immunization schedule, showed the highest affinity (3.20 ± 0.25x108L/mol) and recognized a different epitope to others MAbs. CBSSRBD-S.11 additivity index versus CBSSRBD-S.1, CBSSRBD-S.2, CBSSRBD-S.3, CBSSRBD-S.4, CBSSRBD-S.6, CBSSRBD-S.7 and CBSSRBD-S.8 was 80.79%, 41.31%, 52.42%, 67.25%, 80.07%, 57.72% and 95.67%, respectively (assay cut point of 39.18%).The IC50 of 85.5pM obtained with CBSSRBD-S.11was higher than those reported by Tan et al. (2020), who determined the inhibition of two MAbs in mice with IC50 of 316.2pM and 197.2pM. Consequently, CBSSRBD-S.11 showed the highest inhibition of the binding of RBDr4-HRP and hFc-ACE-2.
Fig. 2.
Monoclonal antibody-mediated blockage of RBDr4-HRP and hFc-ACE-2 binding. Inhibition (%) = (1- A450Inh / A450Max) × 100, where A450Inh means the MAbs inhibition to the binding of RBDr4-HRP and hFc-ACE-2 and A450Max means the A450nm of the conjugate without preincubation with antibodies. Data are the mean of duplicate measurements ± SD.
The three neutralizing MAbs obtained were used for the development and validation of a sVNT by the Analytical Laboratory of the Biomedical Research Division of the Center for Genetic Engineering and Biotechnology, Havana, Cuba (unpublished results). This assay is being used to evaluate clinical trial patients with Cuban vaccines. Routine sVNT uses these neutralizing antibodies as a positive control. Moreover, these neutralizing MAbs could be valuables as biological reagents in researches about structural basis for RBD/ACE-2 binding mechanism, and even in preclinical therapeutic modeling.
3.8. Microneutralization of intact SARS-CoV-2 virus in Vero E6 cell line
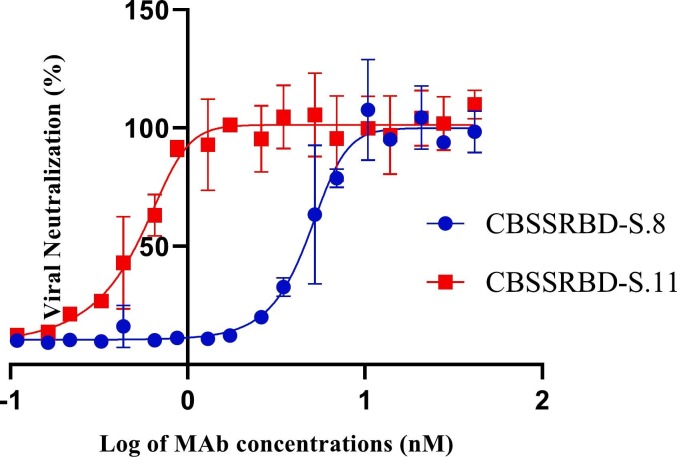
Taking into account the results obtained in the inhibition of the binding of RBD to the ACE-2 receptor by the MAbs CBSSRBD-S.8 and CBSSRBD-S.11, an experiment of neutralization of the infection of the intact virus to Vero E6 cells was carried out. As shown in Fig. 3 , the neutralizing capability of CBSSRBD-S.11 (VN50 = 0.552 nM) was confirmed over CBSSRBD-S.8 (VN50 = 4.85 nM) when D614G strain was used to infect Vero cells. Also CBSSRBD-S.11 neutralized the SARS-CoV-2 strains Alpha (lineage B.1.1.7) and Beta (lineage B.1.351): VN50 = 0.707 nM and VN50 = 0.132 nM, respectively, but not Delta (lineage B.1.617.2), at least in the assayed concentration range (0.0208 nM to 1.33 nM). We do not rule out that at higher concentrations of the CBSSRBD-S.11 antibody, neutralization for Delta strain occurs. In this range, CBSSRBD-S.8 did not show neutralizing activity against none of the strains of concern analyzed.
Fig. 3.
Monoclonal antibody neutralization of SARS-CoV-2 D614G strain. The viral neutralization percentage of MAbs was measured as A540nm sample x 100/ A540nm control. The viral neutralizing at 50% of maximum VN (VN50) was calculated by fitting the curve to a five-parameter logistic regression. Data are the mean of triplicate measurements ± SD.
The potential therapeutic use of those MAbs that showed important neutralizing capacity has not scape to our attention. Even when MAbs have not been yet established as a relevant therapeutic tool for infected patients, the fact that high titer anti-RBD neutralizing antibody response has been considered as a correlate of immunity in vaccine development, suggests that the role of MAbs as therapeutics must be further explored. MAbs characteristics such as epitope specificity and neutralizing capacity, as well as the correct stratifications of patients in relation to time of infection or virus variants, could be important variables to assess in the road to find a place for therapeutics MAbs in COVID-19. It is well known that murine MAbs can generate undesirable immunogenic responses in humans that restrict their use as therapeutics. Humanized murine technology has been applied to derive neutralizing MAbs directed to the RBD-S protein (Chen et al., 2020, Wu et al., 2020). In that sense, the specificity and affinity for RBD-S of our neutralizing MAbs suggest that they could be humanized in order to test their therapeutic potential. Taking into account that these MAbs recognize different epitopes, the combination of them in therapy could be useful to reduce the events of antibody-resistant strains and avoid treatment failure, as explained Taylor et al. (2021).
4. Conclusions
Five of the murine antibodies obtained against the RBD protein of SARS-CoV-2 have affinities in the order of 108L/mol. Six of them show a specific recognition of different recombinant RBDs in solution. A novel procedure for determining the additivity index cut point demonstrates that these anti-RBD antibodies recognize at least five different epitopes. Two MAbs reveal significant neutralization against SARS-CoV-2 in an ACE2-RBD binding inhibition assay and a neutralizing assay with intact SARS-CoV-2 virus. The high affinity MAbs, CBSSRBD-S.8 and CBSSRBD-S.7, recognize different epitopes and are therefore recommended for the development of a sandwich ELISA to quantify recombinant RBD-S antigens in biofabrication processes, in pharmacokinetic studies of preclinical and clinical trials, as well as other SARS-CoV-2 research projects.
Funding
This work was supported with funds from the BioCubaFarma, the Center for Genetic Engineering and Biotechnology, and by the Grant of the National Science and Technology Program- Biotechnology, Pharmaceutical Industry and Medical Technologies of the Ministry of Science and Technology, project code PN385LH007-048. The Civilian Defense Scientific Research Center supported the microneutralization assays.
Authors' contributions
OB, DD, AD performed the generation of hybridomas and cell culture; CH and DA designed and performed the experiments related to the affinity constants and evaluation of the specificity of the MAbs, Additivity test by ELISA, sandwich ELISA for RBDr2 antigen quantification, YL and JP performed the immunization schedule; JP performed the animal care and ascites production; YL purification of MAbs; OV performed the MAbs conjugation, RBD conjugation and Western blot assays; MP determined the MAbs isotypes; CT and LB performed the indirect ELISA for evaluating anti-RBD-S antibodies and SDS-PAGE to evaluate antibodies purity; GL performed the RBDr4-HRP and hFc-ACE-2 binding inhibition by the monoclonal antibodies; IP performed the MAbs conjugation; MB and AF designed and performed the assay of microneutralization of SARS-CoV-2 virus in Vero E6 cell line; CH, DA, AD prepared the draft of the manuscript; YC and EP supervised the study and revision of the manuscript. All authors have approved the final version of the manuscript.
Declaration of Competing Interest
Authors declare no conflict of interest to publishing this information that has not been publishing in any other scientific journal.
Acknowledgments
The authors express thanks to the professors Emilio Carpio and Merardo Pujol, for their assistance in language and writing during the preparation of the manuscript. Also, we are grateful for the technical assistance of Magalis Delgado, Lisbeth Ulloa, Ricardo Pina, María Abreu, Adaleikys Marrero and Javier Díaz. We would like to thank to Belinda Sánchez at the Center for Molecular Immunology and Dagmar García at the Finlay Institute of Vaccines for providing the recombinant RBD spike proteins and thanks to Otto Cruz at National AIDS Research Laboratory (LISIDA) of the Civilian Defense Scientific Research Center for the exchange during viral neutralization experiments.
Data availability
Data will be made available on request.
References
- Andreano E., Piccini G., Licastro D., Casalino L., Johnson N.V., Paciello I., Monego S.D., Pantano E., Manganaro N., Manenti A., Manna R., Casa E., Hyseni I., Benincasa L., Montomoli E., Amaro R.E., McLellan J.S., Rappuoli R. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1101/2020.12.28.424451. 2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- J.David Beatty, Barbara G. Beatty, William G. Vlahos, Measurement of monoclonal antibody affinity by non-competitive enzyme immunoassay, Journal of Immunological Methods, Volume 100, Issues 1–2, 1987, Pages 173-179, ISSN 0022-1759, 10.1016/0022-1759(87)90187-6. [DOI] [PubMed]
- Bertoglio F., Meier D., Langreder N., Steinke S., Rand U., Simonelli L., Heine P.A., Ballmann R., Schneider K.-T., Roth K.D.R., Ruschig M., Riese P., Eschke K., Kim Y., Schäckermann D., Pedotti M., Kuhn P., Zock-Emmenthal S., Wöhrle J., Kilb N., Herz T., Becker M., Grasshoff M., Wenzel E.V., Russo G., Kröger A., Brunotte L., Ludwig S., Fühner V., Krämer S.D., Dübel S., Varani L., Roth G., Čičin-Šain L., Schubert M., Hust M. SARS-CoV-2 neutralizing human recombinant antibodies selected from pre-pandemic healthy donors binding at RBD-ACE2 interface. Nat. Commun. 2021;12:1577. doi: 10.1038/s41467-021-21609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman A.P., Tang X., Lee J.R., Chida A., Mercer K., Wharton R.E., Kainulainen M., Harcourt J.L., Martines R.B., Schroeder M., Zhao L., Bryksin A., Zhou B., Bergeron E., Bollweg B.C., Tamin A., Thornburg N., Wentworth D.E., Petway D., Bagarozzi D.A., Finn M.G., Goldstein J.M. Rapid development of neutralizing and diagnostic SARS-COV-2 mouse monoclonal antibodies. Sci. Rep. 2021;11:9682. doi: 10.1038/s41598-021-88809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Purcell L.A., Snell G., Veesler D. Tackling COVID-19 with neutralizing monoclonal antibodies. Cell. 2021;184:3086–3108. doi: 10.1016/j.cell.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSilva B., Smith W., Weiner R., Kelley M., Smolec J., Lee B., Khan M., Tacey R., Hill H., Celniker A. Recommendations for the bioanalytical method validation of ligand-binding assays to support pharmacokinetic assessments of macromolecules. Pharm. Res. 2003;20:1885–1900. doi: 10.1023/b:pham.0000003390.51761.3d. [DOI] [PubMed] [Google Scholar]
- Friguet B., Djavadi-Ohaniance L., Pages J., Bussard A., Goldberg M. A convenient enzyme-linked immunosorbent assay for testing whether monoclonal antibodies recognize the same antigenic site. Application to hybridomas specific for the beta 2-subunit of Escherichia coli tryptophan synthase. J. Immunol. Methods. 1983;60:351–358. doi: 10.1016/0022-1759(83)90292-2. [DOI] [PubMed] [Google Scholar]
- Galfrè G., Milstein C. Methods in Enzymology, Immunochemical Techniques: Part B. Academic Press; 1981. [1] preparation of monoclonal antibodies: strategies and procedures; pp. 3–46. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J., Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOV.UK . GOV.UK; 2021. COVID-19 (SARS-CoV-2) Variants [WWW Document]https://www.gov.uk/government/collections/new-sars-cov-2-variant URL. (accessed 5.18.21) [Google Scholar]
- GOV.UK . GOV.UK; 2021. Investigation of SARS-CoV-2 Variants of Concern: Technical Briefings [WWW Document]https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201 (accessed 5.18.21) [Google Scholar]
- Guo Y., Kawaguchi A., Takeshita M., Sekiya T., Hirohama M., Yamashita A., Siomi H., Murano K. Potent mouse monoclonal antibodies that block SARS-CoV-2 infection. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggarty A., Legler C., Krantz M.J., Fuks A. Epitopes of carcinoembryonic antigen defined by monoclonal antibodies prepared from mice immunized with purified carcinoembryonic antigen or HCT-8R cells. Cancer Res. 1986;46:300–309. [PubMed] [Google Scholar]
- He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M., Jiang S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004;324:773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-Y., Lee J.-H., Kim M.J., Park S.C., Choi M., Lee W., Ku K.B., Kim B.T., Changkyun Park E., Kim H.G., Kim S.I. Development of a SARS-CoV-2-specific biosensor for antigen detection using scFv-Fc fusion proteins. Biosens. Bioelectron. 2021;175 doi: 10.1016/j.bios.2020.112868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakidis N.C., López-Cortés A., González E.V., Grimaldos A.B., Prado E.O. SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. Npj Vaccin. 2021;6:28. doi: 10.1038/s41541-021-00292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Choi M., Jung Y., Lee S.K., Lee C.-S., Kim Jung, Kim Jongwoo, Kim N.H., Kim B.-T., Kim H.G. A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2 (ACE2) Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limonta-Fernández M., Chinea-Santiago G., Martín-Dunn A.M., Gonzalez-Roche D., Bequet-Romero M., Marquez-Perera G., González-Moya I., Canaan-Haden-Ayala C., Cabrales-Rico A., Espinosa-Rodríguez L.A., Ramos-Gómez Y., Andujar-Martínez I., González-López L.J., de la Iglesia M.P., Zamora-Sanchez J., Cruz-Sui O., Lemos-Pérez G., Cabrera-Herrera G., Valdes-Hernández J., Martinez-Diaz E., Pimentel-Vazquez E., Ayala-Avila M., Guillén-Nieto G. The SARS-CoV-2 receptor-binding domain expressed in Pichia pastoris as a candidate vaccine antigen. medRxiv. 2021 doi: 10.1101/2021.06.29.21259605. 2021.06.29.21259605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ding Y., Zhang J., Chen H., Zhu X., Cai X., Liu X., Xie Q. Simple method of monoclonal antibody production against mammalian cellular prion protein. Hybrid. 2010;2005(29):37–43. doi: 10.1089/hyb.2009.0058. [DOI] [PubMed] [Google Scholar]
- Makdasi E., Levy Y., Alcalay R., Noy-Porat T., Zahavy E., Mechaly A., Epstein E., Peretz E., Cohen H., Bar-On L., Chitlaru T., Cohen O., Glinert I., Achdout H., Israely T., Rosenfeld R., Mazor O. Neutralizing monoclonal anti-SARS-CoV-2 antibodies isolated from immunized rabbits define novel vulnerable spike-protein epitope. Viruses. 2021;13:566. doi: 10.3390/v13040566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandolesi M., Sheward D.J., Hanke L., Ma J., Pushparaj P., Vidakovics L.P., Kim C., Loré K., Dopico X.C., Coquet J.M., McInerney G., Hedestam G.B.K., Murrell B. SARS-CoV-2 protein subunit vaccination elicits potent neutralizing antibody responses. bioRxiv. 2020 doi: 10.1101/2020.07.31.228486. 2020.07.31.228486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marovich M., Mascola J.R., Cohen M.S. Monoclonal antibodies for prevention and treatment of COVID-19. JAMA. 2020;324:131–132. doi: 10.1001/jama.2020.10245. [DOI] [PubMed] [Google Scholar]
- Mire-Sluis A.R., Barrett Y.C., Devanarayan V., Koren E., Liu H., Maia M., Parish T., Scott G., Shankar G., Shores E., Swanson S.J., Taniguchi G., Wierda D., Zuckerman L.A. Recommendations for the design and optimization of immunoassays used in the detection of host antibodies against biotechnology products. J. Immunol. Methods. 2004;289:1–16. doi: 10.1016/j.jim.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Perera R.A.P.M., Ko R., Tsang O.T.Y., Hui D.S.C., Kwan M.Y.M., Brackman C.J., To E.M.W., Yen H., Leung K., Cheng S.M.S., Chan K.H., Chan K.C.K., Li K.-C., Saif L., Barrs V.R., Wu J.T., Sit T.H.C., Poon L.L.M., Peiris M. Evaluation of a SARS-CoV-2 surrogate virus neutralization test for detection of antibody in human, canine, cat, and Hamster sera. J. Clin. Microbiol. 2020;59 doi: 10.1128/JCM.02504-20. /jcm/59/2/JCM.02504-20.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Ke B., Feng Q., Yang D., Lian Q., Li Z., Lu L., Ke C., Liu Z., Liao G. Construction and immunogenic studies of a mFc fusion receptor binding domain (RBD) of spike protein as a subunit vaccine against SARS-CoV-2 infection. Chem. Commun. 2020;56:8683–8686. doi: 10.1039/D0CC03263H. [DOI] [PubMed] [Google Scholar]
- Rattanapisit K., Shanmugaraj B., Manopwisedjaroen S., Purwono P.B., Siriwattananon K., Khorattanakulchai N., Hanittinan O., Boonyayothin W., Thitithanyanont A., Smith D.R., Phoolcharoen W. Rapid production of SARS-CoV-2 receptor binding domain (RBD) and spike specific monoclonal antibody CR3022 in Nicotiana benthamiana. Sci. Rep. 2020;10:17698. doi: 10.1038/s41598-020-74904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I.-C., Tiu C., Hu Z., Chen V.C.-W., Young B.E., Sia W.R., Tan Y.-J., Foo R., Yi Y., Lye D.C., Anderson D.E., Wang L.-F. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat. Biotechnol. 2020;38:1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- Tan H.-X., Juno J.A., Lee W.S., Barber-Axthelm I., Kelly H.G., Wragg K.M., Esterbauer R., Amarasena T., Mordant F.L., Subbarao K., Kent S.J., Wheatley A.K. Immunogenicity of prime-boost protein subunit vaccine strategies against SARS-CoV-2 in mice and macaques. Nat. Commun. 2021;12:1403. doi: 10.1038/s41467-021-21665-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P.C., Adams A.C., Hufford M.M., de la Torre I., Winthrop K., Gottlieb R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021:1–12. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UN News . Not. ONU; 2020. 2020: el año de la pandemia de COVID-19 que cerró el mundo [WWW Document]https://news.un.org/es/story/2020/12/1486082 (accessed 5.13.21) [Google Scholar]
- Valdés Y., Santana D., Paquet F., Fernández S., Climent Y., Chiodo F., Rodríguez Noda L., Ramirez B., Leon K., Hernandez T., Castellanos-Serra L., Garrido R., Chen G.-W., Rivera Dagmar, Rivera Daniel, Verez-Bencomo V. Molecular aspects concerning the use of the SARS-CoV-2 receptor binding domain as a target for preventive vaccines. ACS Cent. Sci. 2021 doi: 10.1021/acscentsci.1c00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes-Balbin Y., Santana-Mederos D., Quintero L., Fernández S., Rodriguez L., Sanchez Ramirez B., Perez-Nicado R., Acosta C., Méndez Y., Ricardo M.G., Hernandez T., Bergado G., Pi F., Valdes A., Carmenate T., Ramirez U., Oliva R., Soubal J.-P., Garrido R., Cardoso F., Landys M., Gonzalez H., Farinas M., Enriquez J., Noa E., Suarez A., Fang C., Espinosa L.A., Ramos Y., González L.J., Climent Y., Rojas G., Relova-Hernández E., Cabrera Infante Y., Losada S.L., Boggiano T., Ojito E., León K., Chiodo F., Paquet F., Chen G.-W., Rivera D.G., Garcia-Rivera D., Verez Bencomo V. SARS-CoV-2 RBD-tetanus toxoid conjugate vaccine induces a strong neutralizing immunity in preclinical studies. ACS Chem. Biol. 2021 doi: 10.1021/acschembio.1c00272. [DOI] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO WHO Director-General's Opening Remarks at the Media Briefing on COVID-19 - 11 March 2020 [WWW Document] 2020. https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020
- WHO WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard With Vaccination Data [WWW Document] 2021. https://covid19.who.int/ (accessed 7.1.21)
- WHO COVID-19 Vaccines [WWW Document] 2021. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/covid-19-vaccines (accessed 5.12.21)
- WHO Draft Landscape and Tracker of COVID-19 Candidate Vaccines [WWW Document] 2021. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed 5.12.21)
- Wilson M.B., Nakane P.K. Immunofluorescence and Related Staining Techniques. Elsevier/North Holland Biomedical Press; 1978. Recent development in the periodate method of conjugating horseradish peroxidase(HRPO) to antibodies; pp. 215–224. [Google Scholar]
- Wu, F., Wang, A., Liu, M., Wang, Q., Chen, J., Xia, S., Ling, Y., Zhang, Y., Xun, J., Lu, L., Jiang, S., Lu, H., Wen, Y., Huang, J., 2020. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv 2020.03.30.20047365. 10.1101/2020.03.30.20047365. [DOI]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Wang R., Ju B., Yu J., Zhang Y., Liu N., Wang J., Zhang Q., Chen P., Zhou B., Li Y., Shen Y., Zhang S., Tian L., Guo Y., Xia L., Zhong X., Cheng L., Ge X., Zhao J., Wang H.-W., Wang X., Zhang Z., Zhang L., Zhou Q. Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies. Cell Res. 2021;31:517–525. doi: 10.1038/s41422-021-00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wan Y., Liu C., Li Y., Liu Z., Lin C., Chen J. Identification and characterization of a monoclonal antibody blocking the SARS-CoV-2 spike protein–ACE2 interaction. Cell. Mol. Immunol. 2021;1–3 doi: 10.1038/s41423-021-00684-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zai J., Yi K., Xie L., Zhu J., Feng X., Li Y. Dual monoclonal antibody-based sandwich ELISA for detection of in vitro packaged Ebola virus. Diagn. Pathol. 2018;13:96. doi: 10.1186/s13000-018-0773-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhartchouk A.N., Sharon C., Satkunarajah M., Auperin T., Viswanathan S., Mutwiri G., Petric M., See R.H., Brunham R.C., Finlay B.B., Cameron C., Kelvin D.J., Cochrane A., Rini J.M., Babiuk L.A. Immunogenicity of a receptor-binding domain of SARS coronavirus spike protein in mice: implications for a subunit vaccine. Vaccine. 2007;25:136–143. doi: 10.1016/j.vaccine.2006.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Davis B.D., Chen S.S., Martinez J.M.S., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 strain in Southern California, USA. medRxiv. 2021 doi: 10.1101/2021.01.18.21249786. 2021.01.18.21249786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Liu Q., Du L., Lu L., Jiang S. Receptor-binding domain as a target for developing SARS vaccines. J. Thorac. Dis. 2013;5(Suppl. 2):S142–S148. doi: 10.3978/j.issn.2072-1439.2013.06.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.