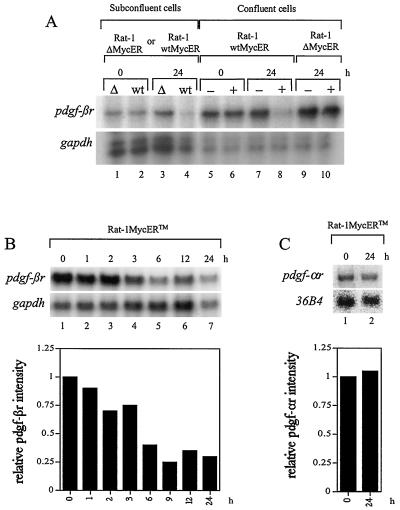
FIG. 3.
pdgf-βr mRNA expression is suppressed in response to Myc induction. (A) pdgf-βr mRNA expression in Rat-1 wtMycER (wt) and Rat-1 ΔMycER (Δ) cells was analyzed under both subconfluent and confluent conditions. Cell lines were grown either as asynchronous subconfluent populations and treated with β-estradiol (lanes 1 to 4) or as confluent quiescent cultures and treated with either ethanol as control (−) or β-estradiol (+) (lanes 5 to 10). At the indicated intervals after β-estradiol treatment, RNA was extracted and total RNA (10 μg) was analyzed by the RNase protection assay to detect the expression of endogenous pdgf-βr and control gapdh genes. (B) Confluent Rat-1 wtMycERTM cells were maintained in low serum (0.1% FBS–αMEM) for 2 days and then exposed to 100 nM OH-T to induce exogenous MycERTM activity. At the indicated intervals after OH-T treatment, RNA was extracted and 10 μg of total RNA was analyzed by the RNase protection assay using single-stranded probes complementary to endogenous pdgf-βr- and gapdh-specific sequences. The histogram shows quantitation of the pdgf-βr signal normalized to gapdh, as determined by densitometry. (C) Northern blot analysis of pdgf-αr and 36B4 expression in OH-T-stimulated Rat-1 wtMycERTM cells. Confluent quiescent Rat-1 wtMycERTM cells were exposed to 100 nM OH-T to induce exogenous MycERTM activity. At the indicated intervals after OH-T treatment, RNA was extracted and 10 μg of total RNA was analyzed by Northern blotting using gene-specific probes for endogenous pdgf-αr- and 36B4-specific sequences. The plotted data show the pdgf-αr signal as normalized to 36B4, as determined by densitometry.