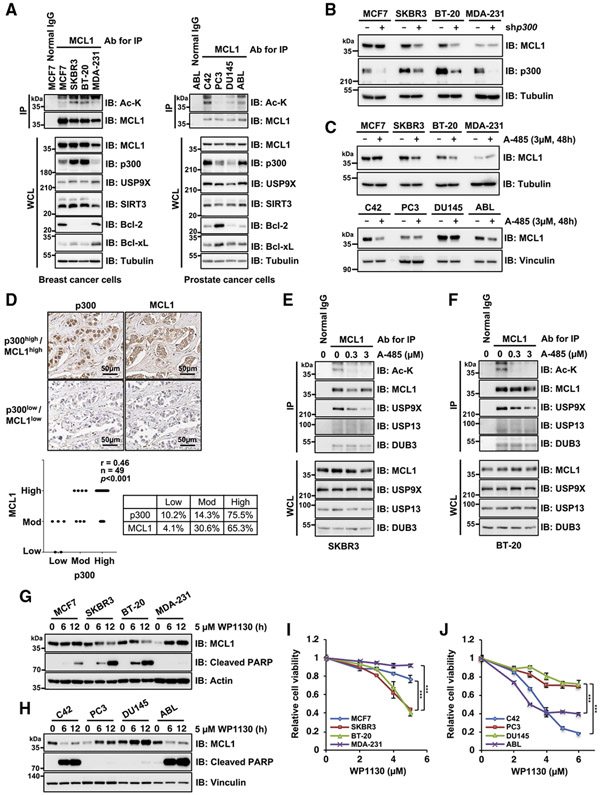
Figure 6. MCL1 acetylation promotes cancer cell survival in a p300- and USP9X-dependent manner.
(A) MCL1 acetylation levels correlate with p300 expression in breast and prostate cancer cell lines. Shown is IB analysis of WCLs and anti-MCL1 immunoprecipitates derived from a panel of breast and prostate cancer cell lines.
(B) p300 depletion impairs MCL1 protein abundance in breast cancer cells with high p300 and acetylated MCL1 levels. Shown is IB analysis of WCLs derived from breast cancer cell lines stably expressing the lentiviral shRNA specific for GFP or p300.
(C) Treatment with the p300/CBP inhibitor A-485 results in decreased MCL1 protein levels in breast and prostate cancer cells with high p300 and acetylated MCL1 levels. Shown is IB analysis of WCLs derived from breast and prostate cancer cell lines treated with A-485 (3 μM) for 48 h before harvesting.
(D) Representative images of MCL1 and p300 expression in breast tumor cells as assessed by immunohistochemistry (IHC). MCL1 and p300 levels were classified as low, moderate, or high, based on the intensities of the IHC staining, and a Spearman correlation test was conducted. Scale bar, 50 μm. See also Table S1.)
(E and F) Treatment with the p300/CBP inhibitor A-485 reduces MCL1 acetylation and promotes dissociation of USP9X from MCL1. Shown is IB analysis of WCLs and anti-MCL1 immunoprecipitates derived from SKBR3 (E) and BT-20 (F) treated overnight with the indicated concentrations of A-485 before harvesting.
(G and H) The USP9X inhibitor WP1130 effectively induces activation of the apoptotic pathway in cells with higher levels of acetylated MCL1. Shown is IB analysis of WCLs derived from the indicated breast cancer (G) and prostate cancer (H) cell lines. These cells were treated with WP1130 (5 μM) for the indicated periods before harvesting.
(I and J) High acetylated MCL1 levels correlate with increased sensitivity to WP1130 in breast and prostate cancer cell lines. Breast cancer (I) and prostate cancer (J) cell lines were treated with the indicated concentrations of WP1130 for 24 h and subjected to cell viability assays. Data are presented as mean ± SD; n = 3 biological replicates; ***p < 0.001. Data in (A)–(C) and (E)–(H) are representative of at least two independent experiments. See also Figure S6.