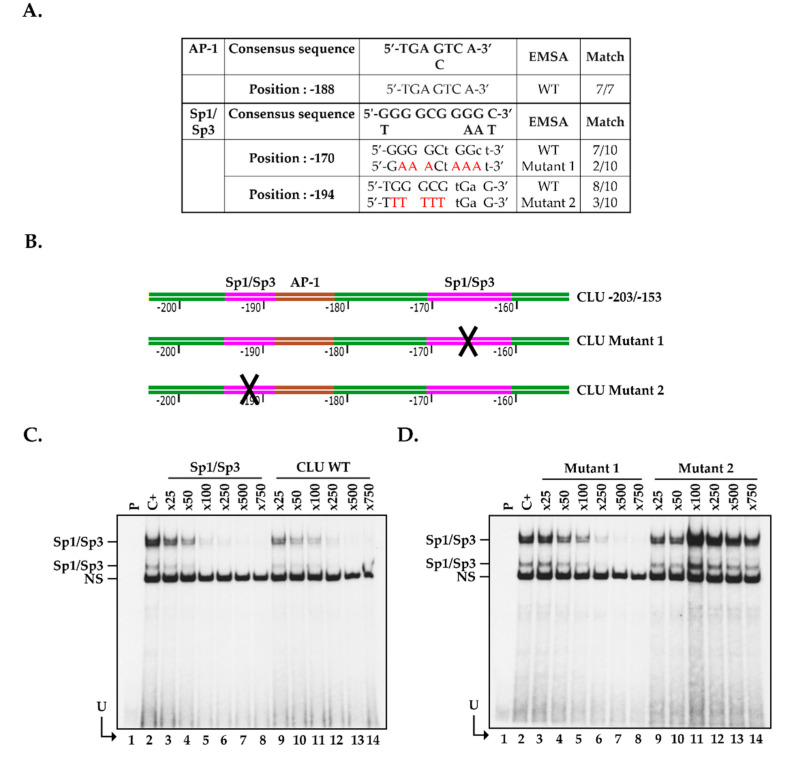
Figure 5.
Sp1/Sp3 binding sites in the -203/-153 CLU promoter segment. (A) DNA sequence of the AP-1 and Sp1/Sp3 target sites from the CLU promoter identified at position -170, -188 and -194, respectively, aligned with their corresponding prototypical sequence. The nucleotides preserved between the CLU TF sites and their consensus sequences are shown in capital letters along with those selected for site-directed mutation (in red). (B) Schematic representation of the -203/-153 CLU promoter segment used in EMSA, and its derivatives that bear mutations into either the -170 (CLU Mutant 1) or the -194 (CLU Mutant 2) Sp1 site (the mutated site is indicated by an ‘X’). (C) Nuclear proteins (15 µg) from control hCECs (Epi 70x) were incubated with the Sp1/Sp3-labeled probe bearing the consensus sequence for the Sp1/Sp3 TFs either alone (C+) or in the presence of increasing molar excesses (25 to 750-fold) of unlabeled CLU-203/-153, or of competitors bearing the target site for Sp1/Sp3 and run at 110V for 6.5 h. P: labeled probe without added proteins, U: free probe. (D) Same as in panel (C), except that the Sp1/Sp3-labeled probe was incubated either alone (C+) or in the presence of the unlabeled CLU Mutant 1 or CLU Mutant 2 oligomers.