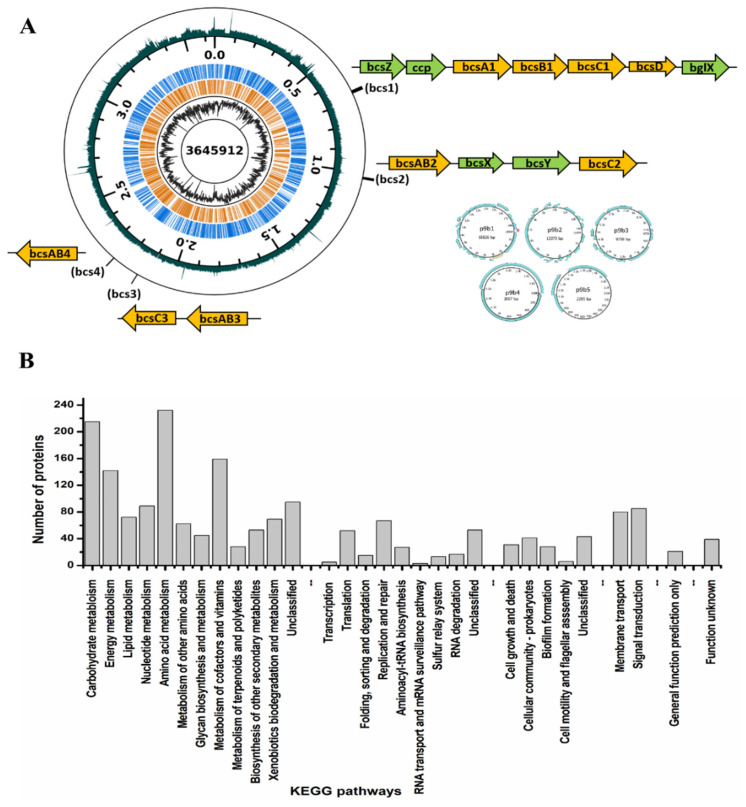
Figure 4.
Overview of K. rhaeticus ENS9b genome. (A) Genome map. The K. rhaeticus ENS9b genome totals 3.69 Mbp (GC% of 63.05%), containing 3360 protein-coding gene predictions, 3 rRNAs, 1 tmRNA, 46 tRNAs and 3 non coding RNAs. A genome completeness of 100% was identified from CheckM. The genome consists of a chromosome of 3.65 Mbp (3302 predicted protein-coding regions). At least three putative plasmids; p9b1 (18.8 Kbp), p9b2 (13.4 Kbp), p9b3 (9.8 Kbp), were identified in K. rhaeticus ENS9b genome ((A) inset). Although the replicon could not be determined, as similarly reported for other K. rhaeticus isolates [7,19], two putative plasmid sequences of sizes 3 Kbp (p9b4) and 2.3 Kbp (p9b5) were detected by plasmidSPAdes. Seven contigs (totaling 90 Kbp) could not be confidently placed in the genome. The displayed data from center to perimeter are: chromosome size (bp), GC-percentage, CDSs on the reverse strand (in the middle, orange), CDSs on the forward strand (in the middle, blue), chromosome position (major ticks 500 kbp, minor ticks 100 kbp), and coverage (outermost). Both GC-percentage and coverage were calculated with 1 kbp window size. GC-percentage ranged from 33.6% to 75.5% and coverage (number of reads mapping to the locus) from 380 to 49600. Coverage is displayed with logarithmic scaling. The bcs operon (orange filled arrows) and the flanking accessory genes (green filled arrows) indicated in the manually curated figures directs to their position in the genome. (B) KEGG classification and functional categorization.