Figure 1.
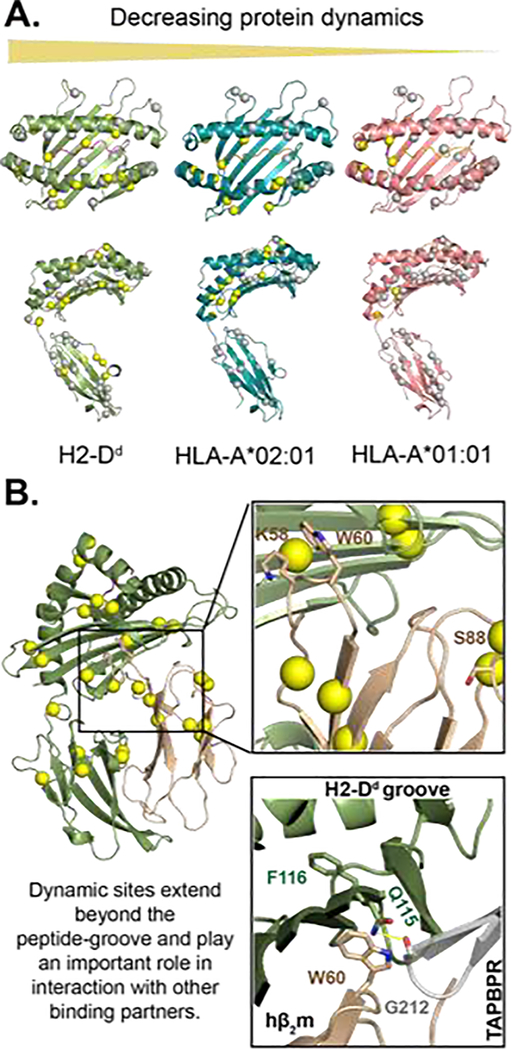
MHC-I exhibits allele-specific conformational dynamics that influence its interaction with its binding partners. A) microseconds-to-milliseconds dynamics profiles of three MHC-I molecules in decreasing order of flexibility from left to right. Conformational flexibility of methyl probes was measured through 13C single-quantum Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion NMR experiments by McShan et al. (2019). The spheres represent methyl groups of Ala, Ile, Leu, or Val on the structure of H2-Dd/P18-I10 (PDB: 3ECB), HLA-A*02:01/TAX (PDB: 1DUZ), and HLA-A*01:01/NRASQ61K (PDB: 6MPP). Methyl sites with increased dynamics are shown in yellow, and areas not exhibiting dynamics are shown in gray. B) Protein dynamics extend beyond the peptide-binding groove to the distal α3 and hβ2m molecules. Similar to figure 1 A, the yellow spheres are methyl groups exhibiting dynamics [9]. Non-methyl dynamics (K58, W60, and S88) on the nanosecond timescale have previously been reported for hβ2m bound to HLA-B*27:09 [23, 26]. Residues K58 and S88 interact with T cell CD8 co-receptor and NK cell receptor Ly49A [26]. NMR chemical shift perturbations analysis revealed micro polymorphism and peptide-occupancy modulate W60 conformational heterogeneity [24]. The proximity of W60, the peptide-binding groove, and the TAPBPR hairpin loop (residue 210–213) suggest that TAPBPR could potentially sense peptide occupancy through hβ2m.