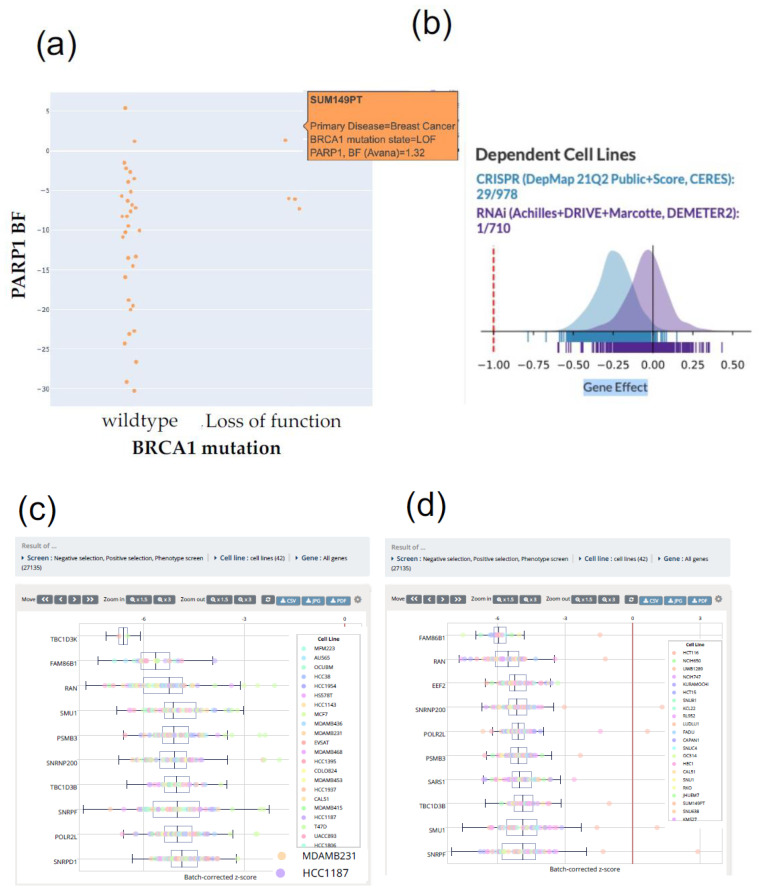
Figure 2.
CRISPR library database. (a) Result of BRCA1 mutation and PARP1 BF from PICKLES (http://pickles.hart-lab.org (accessed on 24 October 2021)). BRCA loss-of-function cell lines are dependent on PARP1 compared with BRCA wild-type cell lines. BF: Bayes factor (b) PARP1-dependent cell line from Depmap portal (https://depmap.org/portal/ (accessed on 24 October 2021)). Depmap’s “Dependent Cell Lines” category suggests that a chosen gene is a ‘common essential’ or ‘strongly selective’ judgment for cell lines regarding CRISPR and RNAi database. Because searching ‘PARP1′ in Depmap showed no judgment in the “Dependent Cell Lines” category, PARP1 is not an essential gene for most cell lines and not-selective drug targets. (c) Top essential genes of the breast cancer cell lines from iCSDB (https://www.kobic.re.kr/icsdb/ (accessed on 24 October 2021)). The result of the top 10 essential genes of the 42 breast cancer cell lines in iCSDB. The genes essentiality score is depicted with a boxplot. Colored dot points each cell line, annotated in square right side (for example: orange dot MDAMB231 cell line, purple dot HCC1187 cell line). (d) Top essential genes of the BRCA1/2 deleted cell lines from iCSDB (https://www.kobic.re.kr/icsdb/ (accessed on 24 October 2021)). The result of top 10 essential genes of the 42 BRCA1/2 deleted cell lines (not organ-specific) in iCSDB. The genes essentiality score is depicted with a boxplot. Colored dot points each cell line, annotated in square right side.