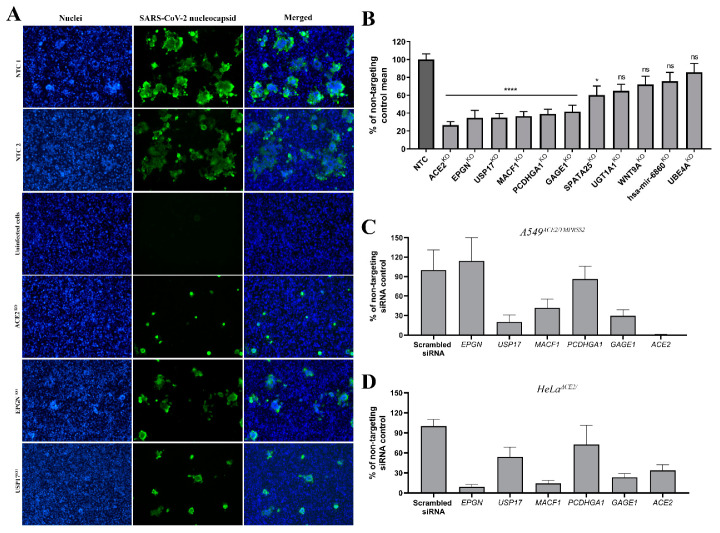
Figure 4.
Identification of essential factors for SARS-CoV-2 replication in modified HeLaACE2 and A549ACE2/TMPRSS2 cells. (A) Immunostaining assay was performed as described in Section 2; cells were fixed after 24 hours pi. Blue color denotes nuclei, green SARS-CoV-2 nucleocapsid protein. (B) SARS-CoV-2 replication was evaluated in modified HeLaACE2 cells transduced with vectors harboring the template for sgRNA knockouts of ACE2, EPGN, USP17, MACF1, PCDHGA1, GAGE1, SPATA25, UGT1A1, WTN9A, hsa-mir-6860, and UBE4A genes. Two sgRNAs non-targeting any sequence in the genome were used as a control (NTC). Inhibition of viral infection was assessed 48 hours pi. by RT-qPCR, data were normalized and presented as percentage of the non-targeting control mean. Data are presented as a mean ± SEM from three independent experiments, each performed in triplicate or quadruplicate. Data were analyzed with Shapiro–Wilk and Brown-Forsythe tests. To determine the significance of differences between compared means, one-way ANOVA with post hoc Dunnett’s test was used. Values statistically significant are indicated by asterisks: * p < 0.05, **** p < 0.0001, ns—non-significant. SARS-CoV-2 replication was evaluated in modified A549ACE2/TMPRSS2 (C) and HeLaACE2 (D) cells transfected with siRNA targeting ACE2, EPGN, USP17, MACF1, PCDHGA1, and GAGE1 genes. Non-targeting siRNA was used as a control (NTC). Inhibition of viral infection was assessed 48 hpi by RT-qPCR, data were normalized and presented as percentage of the non-targeting siRNA control. Data are presented as a mean ± SEM from three independent experiments, each performed in triplicate or quadruplicate.